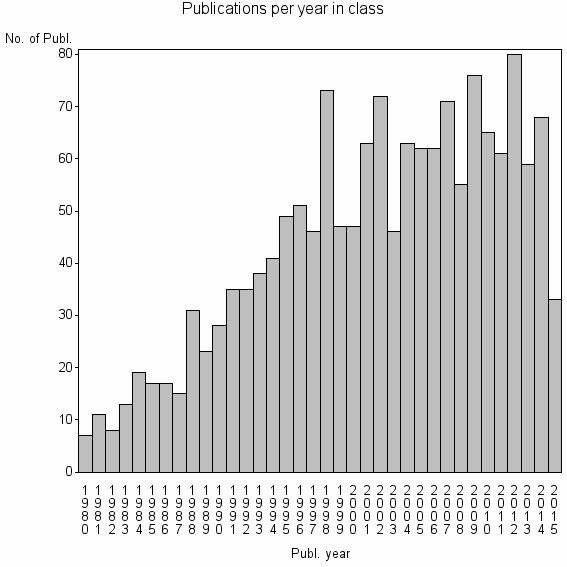
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 5219 | 1587 | 41.1 | 81% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 302 | 17871 | TRANSLESION SYNTHESIS//DNA POLYMERASE//RECA PROTEIN |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | DNA POLYMERASE | Author keyword | 27 | 16% | 9% | 149 |
| 2 | DNA POLYMERASE BETA | Author keyword | 20 | 28% | 4% | 60 |
| 3 | DNA POLYMERASE LAMBDA | Author keyword | 18 | 55% | 1% | 22 |
| 4 | DNA POLYMERASE MU | Author keyword | 11 | 100% | 0% | 6 |
| 5 | DNA POLYMERASE I | Author keyword | 6 | 31% | 1% | 17 |
| 6 | THERMOSTABLE DNA POLYMERASE | Author keyword | 6 | 47% | 1% | 9 |
| 7 | FAMILY X | Author keyword | 6 | 100% | 0% | 4 |
| 8 | EXONUCLEASE ACTIVITY | Author keyword | 5 | 47% | 1% | 8 |
| 9 | BASS MOL STRUCT BIOL | Address | 5 | 50% | 0% | 7 |
| 10 | 3 5 EXONUCLEASE | Author keyword | 4 | 36% | 1% | 10 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA POLYMERASE | 27 | 16% | 9% | 149 | Search DNA+POLYMERASE | Search DNA+POLYMERASE |
| 2 | DNA POLYMERASE BETA | 20 | 28% | 4% | 60 | Search DNA+POLYMERASE+BETA | Search DNA+POLYMERASE+BETA |
| 3 | DNA POLYMERASE LAMBDA | 18 | 55% | 1% | 22 | Search DNA+POLYMERASE+LAMBDA | Search DNA+POLYMERASE+LAMBDA |
| 4 | DNA POLYMERASE MU | 11 | 100% | 0% | 6 | Search DNA+POLYMERASE+MU | Search DNA+POLYMERASE+MU |
| 5 | DNA POLYMERASE I | 6 | 31% | 1% | 17 | Search DNA+POLYMERASE+I | Search DNA+POLYMERASE+I |
| 6 | THERMOSTABLE DNA POLYMERASE | 6 | 47% | 1% | 9 | Search THERMOSTABLE+DNA+POLYMERASE | Search THERMOSTABLE+DNA+POLYMERASE |
| 7 | FAMILY X | 6 | 100% | 0% | 4 | Search FAMILY+X | Search FAMILY+X |
| 8 | EXONUCLEASE ACTIVITY | 5 | 47% | 1% | 8 | Search EXONUCLEASE+ACTIVITY | Search EXONUCLEASE+ACTIVITY |
| 9 | 3 5 EXONUCLEASE | 4 | 36% | 1% | 10 | Search 3++5++EXONUCLEASE | Search 3++5++EXONUCLEASE |
| 10 | DNA POLYMERASES | 4 | 15% | 2% | 26 | Search DNA+POLYMERASES | Search DNA+POLYMERASES |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NUCLEOTIDE INCORPORATION | 99 | 54% | 8% | 129 |
| 2 | I KLENOW FRAGMENT | 78 | 47% | 8% | 123 |
| 3 | REPLICATION FIDELITY | 77 | 37% | 11% | 169 |
| 4 | KLENOW FRAGMENT | 74 | 39% | 9% | 149 |
| 5 | MUTATOR MUTANT | 52 | 100% | 1% | 18 |
| 6 | INDUCED FIT MECHANISM | 43 | 53% | 4% | 58 |
| 7 | LARGE FRAGMENT | 32 | 48% | 3% | 50 |
| 8 | BACTERIOPHAGE RB69 | 31 | 92% | 1% | 12 |
| 9 | FIDELITY | 30 | 10% | 18% | 280 |
| 10 | POL LAMBDA | 29 | 88% | 1% | 14 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| DNA polymerases: Structural diversity and common mechanisms | 1999 | 479 | 25 | 76% |
| Structure and mechanism of DNA polymerase beta | 2006 | 164 | 152 | 64% |
| DNA replication fidelity | 2000 | 570 | 189 | 44% |
| The X family portrait: Structural insights into biological functions of X family polymerases | 2007 | 67 | 85 | 78% |
| The kinetic and chemical mechanism of high-fidelity DNA polymerases | 2010 | 48 | 54 | 63% |
| DNA polymerase Family X: Function, structure, and cellular roles | 2010 | 43 | 169 | 67% |
| DNA replication fidelity | 2004 | 277 | 52 | 46% |
| Role of induced fit in enzyme specificity: A molecular forward/reverse switch | 2008 | 63 | 21 | 48% |
| Techniques used to study the DNA polymerase reaction pathway | 2010 | 25 | 38 | 79% |
| Active site tightness and substrate fit in DNA replication | 2002 | 248 | 97 | 38% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BASS MOL STRUCT BIOL | 5 | 50% | 0.4% | 7 |
| 2 | GRP ABIL GENET CANC | 4 | 75% | 0.2% | 3 |
| 3 | GRP GENET ABIL CANC | 4 | 75% | 0.2% | 3 |
| 4 | JOSEPH GOTTSTEIN MEM CANC | 3 | 23% | 0.8% | 13 |
| 5 | OHIO STATE BIOPHYS PROGRAM | 3 | 26% | 0.7% | 11 |
| 6 | CELL MOL BIOSCI ICAMB | 3 | 40% | 0.4% | 6 |
| 7 | THER EUT RADIOL GENET | 3 | 32% | 0.4% | 7 |
| 8 | GRP ABILITE GENET CANC | 2 | 67% | 0.1% | 2 |
| 9 | HEDCO MOL BIOL S | 2 | 19% | 0.5% | 8 |
| 10 | HARVARD MIT JOINT HLTH SCI TECHNOL | 1 | 100% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000242385 | TRANSLESION SYNTHESIS//DNA POLYMERASE ETA//DNA DAMAGE TOLERANCE |
| 2 | 0.0000175802 | DNA POLYMERASE ALPHA//DNA SYNTHESOME//DNA POLYMERASE DELTA |
| 3 | 0.0000160431 | CLAMP LOADER//REPLICATION FACTOR C//DNA REPLICAT |
| 4 | 0.0000153794 | METAL MEDIATED BASE PAIRS//METAL MEDIATED BASE PAIR//UNNATURAL BASE PAIR |
| 5 | 0.0000136684 | COOPERAT LIFE SCI//FRONTIER GENOM DRUG DISCOVERY//DEHYDROALTENUSIN |
| 6 | 0.0000119131 | APE1//AP ENDONUCLEASE//APE1 REF 1 |
| 7 | 0.0000091018 | GILEAD IOCB//REVERSIBLE TERMINATOR//ONE POT METHODOLOGY |
| 8 | 0.0000090286 | T7 RNA POLYMERASE//T7 LYSOZYME//PHAGE RNA POLYMERASE |
| 9 | 0.0000085288 | HOT START//TEDA BIOX SYST BIOTECHNOL//MARINE ANTIFOULING ENGN TECHNOL SHANGDONG PRO |
| 10 | 0.0000084197 | BIOL MOL ELADIO VINUELA//PHAGE PHI 29//LINEAR DNA REPLICATION |