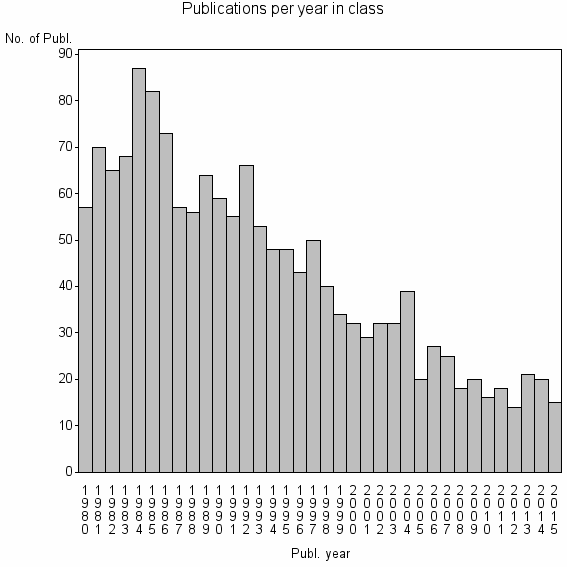
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 5469 | 1553 | 37.4 | 59% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1724 | 6073 | PERILIPIN//LIPID DROPLET//OXYSTEROL BINDING PROTEIN |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CYTIDYLYLTRANSFERASE | Author keyword | 34 | 69% | 2% | 29 |
| 2 | CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE | Author keyword | 31 | 82% | 1% | 18 |
| 3 | CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE | Author keyword | 25 | 70% | 1% | 21 |
| 4 | PHOSPHATIDYLETHANOLAMINE N METHYLTRANSFERASE | Author keyword | 17 | 59% | 1% | 19 |
| 5 | CANADIAN HLTH GRP MOL CELL BIOL LIPIDS | Address | 16 | 51% | 1% | 23 |
| 6 | ETHANOLAMINEPHOSPHOTRANSFERASE | Author keyword | 12 | 86% | 0% | 6 |
| 7 | PCYT2 | Author keyword | 11 | 100% | 0% | 6 |
| 8 | GRP MOL CELL BIOL LIPIDS | Address | 10 | 25% | 2% | 36 |
| 9 | PHOSPHATIDYLETHANOLAMINE METHYLATION | Author keyword | 9 | 83% | 0% | 5 |
| 10 | CHOLINEPHOSPHOTRANSFERASE | Author keyword | 8 | 45% | 1% | 14 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RAT LIVER CTP | 174 | 92% | 4% | 69 |
| 2 | PHOSPHORYLCHOLINE CYTIDYLYLTRANSFERASE | 143 | 100% | 3% | 39 |
| 3 | CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE | 86 | 40% | 11% | 167 |
| 4 | PHOSPHATIDYLCHOLINE BIOSYNTHESIS | 57 | 34% | 9% | 138 |
| 5 | N METHYLTRANSFERASE 2 | 57 | 95% | 1% | 19 |
| 6 | CHOLINE PHOSPHATE CYTIDYLYLTRANSFERASE | 49 | 64% | 3% | 48 |
| 7 | PHOSPHATIDYLCHOLINE SYNTHESIS | 45 | 37% | 6% | 97 |
| 8 | CDP ETHANOLAMINE | 44 | 100% | 1% | 16 |
| 9 | PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE | 44 | 49% | 4% | 66 |
| 10 | PHOSPHOLIPASE C TREATMENT | 41 | 85% | 1% | 22 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Phospholipid biosynthesis in mammalian cells | 2004 | 168 | 178 | 60% |
| The Kennedy Pathway-De Novo Synthesis of Phosphatidylethanolamine and Phosphatidylcholine | 2010 | 86 | 83 | 48% |
| CTP:phosphocholine cytidylyltransferase | 1997 | 151 | 98 | 79% |
| Phosphatidylcholine and choline homeostasis | 2008 | 138 | 73 | 62% |
| Phospholipid methylation in mammals: from biochemistry to physiological function | 2014 | 9 | 71 | 63% |
| Phosphatidylcholine and the CDP-choline cycle | 2013 | 22 | 156 | 69% |
| EUKARYOTIC PHOSPHOLIPID BIOSYNTHESIS | 1995 | 247 | 194 | 36% |
| Structure and function of choline kinase isoforms in mammalian cells | 2004 | 99 | 66 | 52% |
| REGULATION OF THE BIOSYNTHESIS OF TRIACYLGLYCEROL, PHOSPHATIDYLCHOLINE AND PHOSPHATIDYLETHANOLAMINE IN THE LIVER | 1989 | 191 | 173 | 58% |
| Thematic review series: Glycerolipids. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids | 2008 | 138 | 139 | 38% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CANADIAN HLTH GRP MOL CELL BIOL LIPIDS | 16 | 51% | 1.5% | 23 |
| 2 | GRP MOL CELL BIOL LIPIDS | 10 | 25% | 2.3% | 36 |
| 3 | ATLANTIC | 3 | 13% | 1.5% | 23 |
| 4 | HLTH GRP MOL CELL BIOL LIPIDS | 3 | 60% | 0.2% | 3 |
| 5 | HERITAGE MED 328 | 2 | 23% | 0.5% | 8 |
| 6 | LIPID LIPOPROT GRP | 2 | 16% | 0.6% | 10 |
| 7 | CIHR GRP MOL CELL BIOL LIPIDS | 1 | 21% | 0.4% | 6 |
| 8 | HERITAGE MED 332 | 1 | 38% | 0.2% | 3 |
| 9 | AREA BIOL CIENCIAS BIOL CIENCIAS BIOQUIM | 1 | 100% | 0.1% | 2 |
| 10 | MED PHYSIOL EXPTL CARDIOL SECT | 1 | 100% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000191886 | LIPIN//INO2//RUTGERS LIPID |
| 2 | 0.0000110853 | CHOLINE DEFICIENCY//BETAINE//CHOLINE |
| 3 | 0.0000098928 | ONTOGENY REPROD//MOL EVOLUTIONARY PHYSIOL LUNG//HUMAN FETAL LUNG |
| 4 | 0.0000094571 | FATTY ACID REMODELING//TRANSACYLASE//COA INDEPENDENT TRANSACYLASE |
| 5 | 0.0000090005 | AXON MYELIN TRANSFER//MYELIN PHOSPHOINOSITIDES//SIGNAL TRANSDUCTION IN MYELIN |
| 6 | 0.0000086338 | CITICOLINE//CDP CHOLINE//CHOLINE ALPHOSCERATE |
| 7 | 0.0000074652 | PHOSPHATIDATE PHOSPHOHYDROLASE//SN GLYCEROL 3 PHOSPHATE//GLYCEROPHOSPHATE ACYLTRANSFERASE |
| 8 | 0.0000061476 | FAST PADE TRANSFORM//CHOLINE KINASE//CHOLINE KINASE INHIBITORS |
| 9 | 0.0000060681 | PHOSPHOLIPASE D//PHOSPHATIDIC ACID//PHOSPHOLIPASE D2 |
| 10 | 0.0000060325 | PLASMALOGEN//PLASMALOGENS//PLASMENYLCHOLINE |