Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
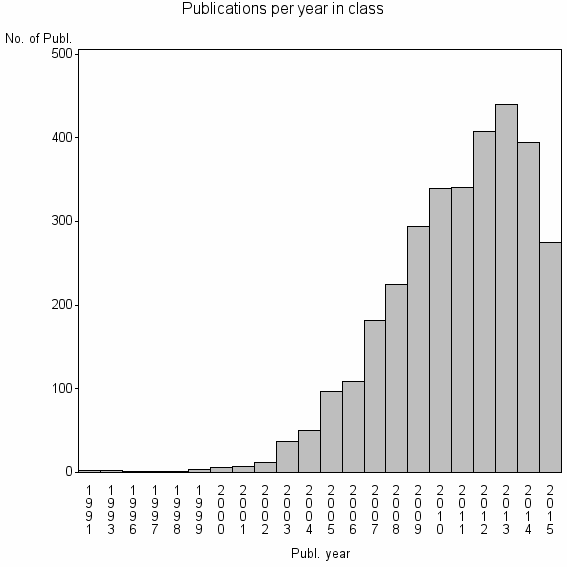
| 563 | 3225 | 52.8 | 93% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | GW182 | Author keyword | 27 | 71% | 1% | 22 |
| 2 | DROSHA | Author keyword | 27 | 41% | 2% | 51 |
| 3 | ARGONAUTE | Author keyword | 23 | 27% | 2% | 73 |
| 4 | LET 7 | Author keyword | 16 | 26% | 2% | 53 |
| 5 | LIN 14 | Author keyword | 15 | 88% | 0% | 7 |
| 6 | ISOMIR | Author keyword | 13 | 58% | 0% | 15 |
| 7 | MIRNP | Author keyword | 12 | 86% | 0% | 6 |
| 8 | SILKWORM MULBERRY GENET IMPROVEMENT | Address | 11 | 60% | 0% | 12 |
| 9 | DICER | Author keyword | 11 | 15% | 2% | 69 |
| 10 | LIN 41 | Author keyword | 11 | 78% | 0% | 7 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GW182 | 27 | 71% | 1% | 22 | Search GW182 | Search GW182 |
| 2 | DROSHA | 27 | 41% | 2% | 51 | Search DROSHA | Search DROSHA |
| 3 | ARGONAUTE | 23 | 27% | 2% | 73 | Search ARGONAUTE | Search ARGONAUTE |
| 4 | LET 7 | 16 | 26% | 2% | 53 | Search LET+7 | Search LET+7 |
| 5 | LIN 14 | 15 | 88% | 0% | 7 | Search LIN+14 | Search LIN+14 |
| 6 | ISOMIR | 13 | 58% | 0% | 15 | Search ISOMIR | Search ISOMIR |
| 7 | MIRNP | 12 | 86% | 0% | 6 | Search MIRNP | Search MIRNP |
| 8 | DICER | 11 | 15% | 2% | 69 | Search DICER | Search DICER |
| 9 | LIN 41 | 11 | 78% | 0% | 7 | Search LIN+41 | Search LIN+41 |
| 10 | TARGET PREDICTION | 10 | 30% | 1% | 29 | Search TARGET+PREDICTION | Search TARGET+PREDICTION |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MICROPROCESSOR COMPLEX | 90 | 39% | 6% | 182 |
| 2 | DROSHA DGCR8 COMPLEX | 67 | 58% | 2% | 77 |
| 3 | LET 7 MICRORNA | 62 | 43% | 3% | 109 |
| 4 | TARGET PREDICTIONS | 53 | 60% | 2% | 58 |
| 5 | MICRORNA TARGETS | 46 | 25% | 5% | 157 |
| 6 | REGULATORY RNA | 45 | 37% | 3% | 99 |
| 7 | MIRNAS | 44 | 20% | 6% | 195 |
| 8 | MAMMALIAN MICRORNAS | 44 | 45% | 2% | 75 |
| 9 | NEGATIVE POSTTRANSCRIPTIONAL REGULATION | 44 | 76% | 1% | 31 |
| 10 | ANIMAL MICRORNAS | 44 | 49% | 2% | 65 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| MicroRNAs: Target Recognition and Regulatory Functions | 2009 | 5235 | 107 | 68% |
| Regulation of microRNA biogenesis | 2014 | 116 | 269 | 52% |
| The complexity of miRNA-mediated repression | 2015 | 13 | 151 | 52% |
| Gene silencing by microRNAs: contributions of translational repression and mRNA decay | 2011 | 623 | 84 | 75% |
| NON-CODING RNA MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship | 2012 | 294 | 109 | 69% |
| Diversifying microRNA sequence and function | 2013 | 154 | 239 | 55% |
| Roles for MicroRNAs in Conferring Robustness to Biological Processes | 2012 | 321 | 87 | 47% |
| The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC | 2012 | 206 | 98 | 69% |
| The widespread regulation of microRNA biogenesis, function and decay | 2010 | 1088 | 171 | 39% |
| Regulation of mRNA Translation and Stability by microRNAs | 2010 | 721 | 181 | 53% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SILKWORM MULBERRY GENET IMPROVEMENT | 11 | 60% | 0.4% | 12 |
| 2 | DIANA | 7 | 67% | 0.2% | 6 |
| 3 | BIOINFORMAT SCI TECHNOL | 4 | 10% | 1.0% | 33 |
| 4 | SIEE | 3 | 100% | 0.1% | 3 |
| 5 | STRATEG TRANSLAT CANC | 3 | 60% | 0.1% | 3 |
| 6 | HENAN INNOVAT ENGN POULTRY GERMPLASM O | 2 | 36% | 0.2% | 5 |
| 7 | BIOL SCI DENT MED PROGRAM | 2 | 67% | 0.1% | 2 |
| 8 | DATABASE PARALLEL COMP HEILONGJIANG PRO | 2 | 67% | 0.1% | 2 |
| 9 | ECOL REMEDIAT SAFE UTILIZAT HEAVY MET P | 2 | 67% | 0.1% | 2 |
| 10 | NANJING DRUM TOWER HOSPMED | 2 | 67% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000241625 | LIN28//LIN28B//LIN28A |
| 2 | 0.0000234354 | O I//VIRAL MICRORNAS//VIRAL MIRNA |
| 3 | 0.0000211260 | JR GRP SFB488//INTERDISZIPLINA ZENTRUM NEUROWISSEN//MIR137 |
| 4 | 0.0000181141 | DICER//DROSHA//MICROPROCESSOR COMPLEX |
| 5 | 0.0000179045 | MARC RUTI BELL VASC BIOL DIS PROGRAM//MIR 33//ABCA1 PROTEIN |
| 6 | 0.0000157277 | MIR 155//MIR 146A//MIRNA 155 |
| 7 | 0.0000155084 | PIRNA//PIWI//PIRNAS |
| 8 | 0.0000145926 | RNA CARDIOVASC//IMTTS//MIR 133 |
| 9 | 0.0000143934 | SMALL ACTIVATING RNA//SARNA//RNA ACTIVATION |
| 10 | 0.0000140841 | MIR156//DCL1//MIR172 |