Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
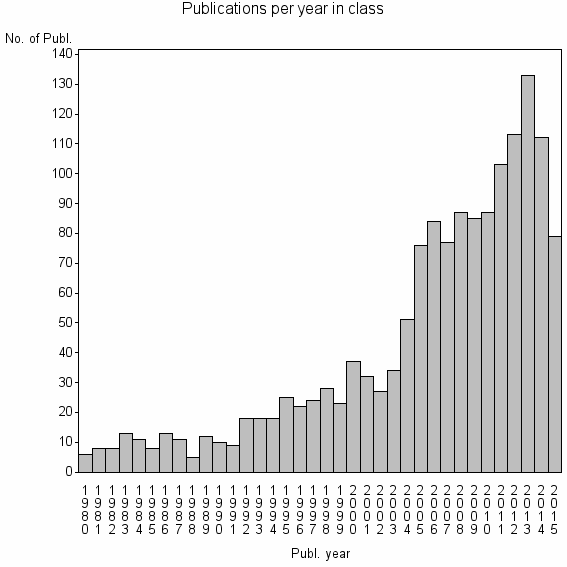
| 5793 | 1507 | 31.9 | 60% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 855 | 10973 | SYSTEMATIC BIOLOGY//SEKT ORNITHOL//FOSSIL BIRDS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PHYLOGENETIC NETWORK | Author keyword | 51 | 61% | 4% | 54 |
| 2 | PHYLOGENETIC NETWORKS | Author keyword | 45 | 64% | 3% | 44 |
| 3 | TIGHT SPAN | Author keyword | 41 | 87% | 1% | 20 |
| 4 | DEEP COALESCENCE | Author keyword | 17 | 72% | 1% | 13 |
| 5 | EVOLUTIONARY TREES | Author keyword | 17 | 50% | 2% | 24 |
| 6 | SUPERTREES | Author keyword | 16 | 60% | 1% | 18 |
| 7 | SYSTEMATIC BIOLOGY | Journal | 16 | 11% | 9% | 139 |
| 8 | ALGORITHMS BIOINFORMAT COMPLEX FORMAL METHODS R | Address | 15 | 67% | 1% | 14 |
| 9 | SPLIT NETWORK | Author keyword | 15 | 88% | 0% | 7 |
| 10 | GENE TREE | Author keyword | 14 | 34% | 2% | 34 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PHYLOGENETIC NETWORK | 51 | 61% | 4% | 54 | Search PHYLOGENETIC+NETWORK | Search PHYLOGENETIC+NETWORK |
| 2 | PHYLOGENETIC NETWORKS | 45 | 64% | 3% | 44 | Search PHYLOGENETIC+NETWORKS | Search PHYLOGENETIC+NETWORKS |
| 3 | TIGHT SPAN | 41 | 87% | 1% | 20 | Search TIGHT+SPAN | Search TIGHT+SPAN |
| 4 | DEEP COALESCENCE | 17 | 72% | 1% | 13 | Search DEEP+COALESCENCE | Search DEEP+COALESCENCE |
| 5 | EVOLUTIONARY TREES | 17 | 50% | 2% | 24 | Search EVOLUTIONARY+TREES | Search EVOLUTIONARY+TREES |
| 6 | SUPERTREES | 16 | 60% | 1% | 18 | Search SUPERTREES | Search SUPERTREES |
| 7 | SPLIT NETWORK | 15 | 88% | 0% | 7 | Search SPLIT+NETWORK | Search SPLIT+NETWORK |
| 8 | GENE TREE | 14 | 34% | 2% | 34 | Search GENE+TREE | Search GENE+TREE |
| 9 | SPECIES TREE | 14 | 26% | 3% | 46 | Search SPECIES+TREE | Search SPECIES+TREE |
| 10 | AGREEMENT FOREST | 14 | 100% | 0% | 7 | Search AGREEMENT+FOREST | Search AGREEMENT+FOREST |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SEQUENCES SUBJECT | 41 | 67% | 2% | 37 |
| 2 | CONSISTENT EVOLUTIONARY HISTORY | 38 | 84% | 1% | 21 |
| 3 | GENE TREES | 35 | 22% | 9% | 141 |
| 4 | ROOTED TRIPLETS | 32 | 85% | 1% | 17 |
| 5 | SPECIES TREES | 32 | 22% | 9% | 131 |
| 6 | SUPERTREES | 26 | 56% | 2% | 32 |
| 7 | RECONCILED TREES | 21 | 52% | 2% | 29 |
| 8 | COPHYLOGENY RECONSTRUCTION PROBLEM | 18 | 89% | 1% | 8 |
| 9 | AGGLOMERATIVE METHOD | 17 | 55% | 1% | 21 |
| 10 | N TREES | 17 | 47% | 2% | 26 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SYSTEMATIC BIOLOGY | 16 | 11% | 9% | 139 |
| 2 | ALGORITHMS FOR MOLECULAR BIOLOGY | 3 | 11% | 2% | 27 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Gene tree discordance, phylogenetic inference and the multispecies coalescent | 2009 | 410 | 63 | 56% |
| Coalescent methods for estimating phylogenetic trees | 2009 | 115 | 66 | 71% |
| Origin of land plants using the multispecies coalescent model | 2013 | 22 | 25 | 36% |
| Computational approaches to species phylogeny inference and gene tree reconciliation | 2013 | 8 | 74 | 70% |
| The evolution of supertrees | 2004 | 147 | 47 | 47% |
| High-Throughput Genomic Data in Systematics and Phylogenetics | 2013 | 28 | 110 | 20% |
| Networks in phylogenetic analysis: new tools for population biology | 2005 | 74 | 51 | 43% |
| Phylogenetic supertrees: assembling the trees of life | 1998 | 226 | 27 | 44% |
| Missing data and the design of phylogenetic analyses | 2006 | 219 | 32 | 31% |
| Phylogenetic inference using whole Genomes | 2008 | 71 | 57 | 25% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ALGORITHMS BIOINFORMAT COMPLEX FORMAL METHODS R | 15 | 67% | 0.9% | 14 |
| 2 | GK STRUKTURBILDUNGSPROZESSE | 8 | 100% | 0.3% | 5 |
| 3 | BIOINFORMAT ZBIT | 8 | 42% | 0.9% | 14 |
| 4 | COMBINATOR GEOMETRY | 5 | 37% | 0.7% | 11 |
| 5 | ISEMUMR 5554 | 4 | 75% | 0.2% | 3 |
| 6 | FSP MATHEMATISIERUNG | 3 | 60% | 0.2% | 3 |
| 7 | MPG PARTNER | 3 | 60% | 0.2% | 3 |
| 8 | DKE | 2 | 44% | 0.3% | 4 |
| 9 | ZBIT | 2 | 23% | 0.5% | 7 |
| 10 | FSPM STRUKTURBILDUNGSPROZESSE | 2 | 33% | 0.3% | 4 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000214898 | CODON MODEL//HETEROTACHY//PHYLOGENETIC INVARIANTS |
| 2 | 0.0000112038 | CIRCOS//BIODADOS//ORTHOLOGY |
| 3 | 0.0000094897 | TREE OF LIFE//HORIZONTAL GENE TRANSFER//LATERAL GENE TRANSFER |
| 4 | 0.0000079033 | PROGRAM COMPUTAT SYST BIOL//PERIODIC SELECTION//CHURINCE |
| 5 | 0.0000075521 | MAPPING BY SEQUENCING//RADSEQ//REDUCED REPRESENTATION LIBRARIES |
| 6 | 0.0000070515 | CULTURAL PHYLOGENETICS//LEXICOSTATISTICS//HUMAN EVOLUTIONARY ECOL GRP |
| 7 | 0.0000068491 | HIERARCHICAL CLASSES//JOURNAL OF CLASSIFICATION//COMBINATORIAL DATA ANALYSIS |
| 8 | 0.0000066827 | CLADISTICS//CLADISTICS-THE INTERNATIONAL JOURNAL OF THE WILLI HENNIG SOCIETY//IDEALISTIC MORPHOLOGY |
| 9 | 0.0000062486 | MACROEVOLUTION//TREE BALANCE//YULE MODEL |
| 10 | 0.0000056036 | MOLECULAR CLOCK//RELAXED CLOCK//PHYLODYNAMICS |