Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
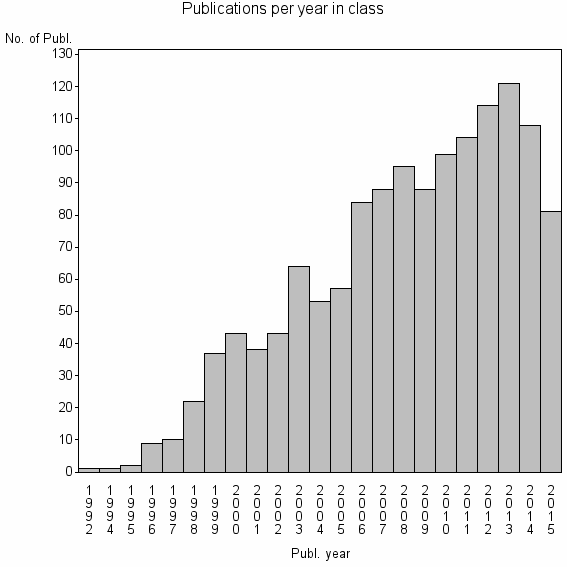
| 6830 | 1362 | 55.6 | 95% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 293 | 18115 | METABOTROPIC GLUTAMATE RECEPTOR//METABOTROPIC GLUTAMATE RECEPTORS//LONG TERM POTENTIATION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | STARGAZIN | Author keyword | 40 | 66% | 3% | 37 |
| 2 | PICK1 | Author keyword | 33 | 60% | 3% | 36 |
| 3 | AMPA RECEPTOR | Author keyword | 19 | 14% | 9% | 123 |
| 4 | AMPA RECEPTORS | Author keyword | 15 | 15% | 7% | 89 |
| 5 | TRANSMEMBRANE AMPA RECEPTOR REGULATORY PROTEIN | Author keyword | 9 | 83% | 0% | 5 |
| 6 | GLUA2 | Author keyword | 9 | 42% | 1% | 16 |
| 7 | TARPS | Author keyword | 8 | 56% | 1% | 10 |
| 8 | SYNAPTIC SCALING | Author keyword | 8 | 32% | 1% | 20 |
| 9 | GLUR1 | Author keyword | 7 | 20% | 2% | 34 |
| 10 | GLUA1 | Author keyword | 7 | 38% | 1% | 15 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | STARGAZIN | 40 | 66% | 3% | 37 | Search STARGAZIN | Search STARGAZIN |
| 2 | PICK1 | 33 | 60% | 3% | 36 | Search PICK1 | Search PICK1 |
| 3 | AMPA RECEPTOR | 19 | 14% | 9% | 123 | Search AMPA+RECEPTOR | Search AMPA+RECEPTOR |
| 4 | AMPA RECEPTORS | 15 | 15% | 7% | 89 | Search AMPA+RECEPTORS | Search AMPA+RECEPTORS |
| 5 | TRANSMEMBRANE AMPA RECEPTOR REGULATORY PROTEIN | 9 | 83% | 0% | 5 | Search TRANSMEMBRANE+AMPA+RECEPTOR+REGULATORY+PROTEIN | Search TRANSMEMBRANE+AMPA+RECEPTOR+REGULATORY+PROTEIN |
| 6 | GLUA2 | 9 | 42% | 1% | 16 | Search GLUA2 | Search GLUA2 |
| 7 | TARPS | 8 | 56% | 1% | 10 | Search TARPS | Search TARPS |
| 8 | SYNAPTIC SCALING | 8 | 32% | 1% | 20 | Search SYNAPTIC+SCALING | Search SYNAPTIC+SCALING |
| 9 | GLUR1 | 7 | 20% | 2% | 34 | Search GLUR1 | Search GLUR1 |
| 10 | GLUA1 | 7 | 38% | 1% | 15 | Search GLUA1 | Search GLUA1 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GLUR1 SUBUNIT | 71 | 55% | 6% | 88 |
| 2 | STARGAZIN | 57 | 71% | 3% | 46 |
| 3 | MOUSE STARGAZER | 46 | 73% | 3% | 35 |
| 4 | 2 DISTINCT MECHANISMS | 40 | 46% | 5% | 65 |
| 5 | EXCITATORY SYNAPSES | 30 | 16% | 12% | 168 |
| 6 | AMPA RECEPTOR TRAFFICKING | 30 | 23% | 9% | 117 |
| 7 | CORNICHON PROTEINS | 30 | 79% | 1% | 19 |
| 8 | AUXILIARY SUBUNITS | 25 | 40% | 4% | 48 |
| 9 | TARP GAMMA 8 | 23 | 100% | 1% | 10 |
| 10 | QUANTAL AMPLITUDE | 22 | 41% | 3% | 41 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| AMPARs and Synaptic Plasticity: The Last 25 Years | 2013 | 70 | 169 | 60% |
| Synaptic AMPA Receptor Plasticity and Behavior | 2009 | 358 | 146 | 44% |
| Homeostatic Control of Presynaptic Neurotransmitter Release | 2015 | 5 | 111 | 26% |
| Assembly of AMPA receptors: mechanisms and regulation | 2015 | 3 | 83 | 35% |
| The Self-Tuning Neuron: Synaptic Scaling of Excitatory Synapses | 2008 | 444 | 95 | 43% |
| The cell biology of synaptic plasticity: AMPA receptor trafficking | 2007 | 414 | 186 | 59% |
| Molecular mechanisms of homeostatic synaptic downscaling | 2014 | 12 | 46 | 63% |
| AMPA receptor trafficking and synaptic plasticity | 2002 | 1377 | 123 | 50% |
| The Expanding Social Network of Ionotropic Glutamate Receptors: TARPs and Other Transmembrane Auxiliary Subunits | 2011 | 109 | 189 | 49% |
| Too Many Cooks? Intrinsic and Synaptic Homeostatic Mechanisms in Cortical Circuit Refinement | 2011 | 164 | 84 | 31% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | UMR 5091 | 5 | 23% | 1.3% | 18 |
| 2 | MRC SYN T PLAST | 4 | 14% | 2.1% | 29 |
| 3 | NANCY PRITZKER | 4 | 17% | 1.7% | 23 |
| 4 | SYN SE NEURAL CIRCUIT UNIT | 3 | 100% | 0.2% | 3 |
| 5 | UR497 | 3 | 100% | 0.2% | 3 |
| 6 | PROGRAM NEUROSCI PHYSIOL | 3 | 40% | 0.4% | 6 |
| 7 | SYN T PLAST | 3 | 12% | 1.5% | 21 |
| 8 | HOWARD HUGHES MED PICOWER LEARNING MEM | 2 | 67% | 0.1% | 2 |
| 9 | NACS PROGRAM | 2 | 67% | 0.1% | 2 |
| 10 | NEUROSCI PHARMACOL HLTH MED SCI | 2 | 67% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000169269 | MAGUK//PDZ DOMAIN//PSD 95 SAP90 |
| 2 | 0.0000163409 | DREBRIN//DENDRITIC SPINE//DENDRITIC SPINES |
| 3 | 0.0000148537 | CYCLOTHIAZIDE//KAINATE RECEPTOR//KAINATE RECEPTORS |
| 4 | 0.0000136803 | KIBRA//PKM ZETA//WWC1 |
| 5 | 0.0000114783 | LONG TERM POTENTIATION//LONG TERM DEPRESSION//DEPOTENTIATION |
| 6 | 0.0000107444 | NEURITIN//ARG31//ACTIVITY REGULATED CYTOSKELETON ASSOCIATED PROTEIN |
| 7 | 0.0000106348 | NR2B//NR2A//NR3A |
| 8 | 0.0000100235 | NEUROLIGIN//SHANK3//NEUREXIN |
| 9 | 0.0000091670 | SYNGAP1//GENES COGNIT PROGRAMME//NEUROCIENCIAS LIM 27 |
| 10 | 0.0000089625 | HOMER PROTEINS//HOMER//HOMER1 |