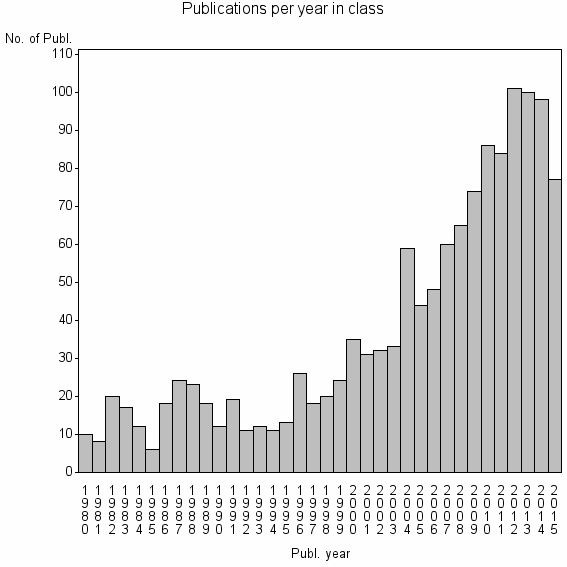
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 6939 | 1349 | 52.9 | 61% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 587 | 13504 | NEW ZEALAND FLORA//NEW ZEALAND JOURNAL OF BOTANY//GENOME SIZE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | POLYPLOIDY | Author keyword | 62 | 16% | 26% | 356 |
| 2 | AUTOPOLYPLOIDY | Author keyword | 22 | 41% | 3% | 41 |
| 3 | ALLOPOLYPLOIDY | Author keyword | 21 | 28% | 5% | 64 |
| 4 | SEQUENCE ELIMINATION | Author keyword | 15 | 88% | 1% | 7 |
| 5 | TRAGOPOGON | Author keyword | 14 | 64% | 1% | 14 |
| 6 | MINORITY CYTOTYPE EXCLUSION | Author keyword | 12 | 86% | 0% | 6 |
| 7 | CYTOGEOGRAPHY | Author keyword | 9 | 38% | 1% | 20 |
| 8 | EXPRESSION LEVEL DOMINANCE | Author keyword | 8 | 100% | 0% | 5 |
| 9 | ALLOPOLYPLOID | Author keyword | 8 | 24% | 2% | 28 |
| 10 | GENOME DOUBLING | Author keyword | 7 | 53% | 1% | 9 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | POLYPLOIDY | 62 | 16% | 26% | 356 | Search POLYPLOIDY | Search POLYPLOIDY |
| 2 | AUTOPOLYPLOIDY | 22 | 41% | 3% | 41 | Search AUTOPOLYPLOIDY | Search AUTOPOLYPLOIDY |
| 3 | ALLOPOLYPLOIDY | 21 | 28% | 5% | 64 | Search ALLOPOLYPLOIDY | Search ALLOPOLYPLOIDY |
| 4 | SEQUENCE ELIMINATION | 15 | 88% | 1% | 7 | Search SEQUENCE+ELIMINATION | Search SEQUENCE+ELIMINATION |
| 5 | TRAGOPOGON | 14 | 64% | 1% | 14 | Search TRAGOPOGON | Search TRAGOPOGON |
| 6 | MINORITY CYTOTYPE EXCLUSION | 12 | 86% | 0% | 6 | Search MINORITY+CYTOTYPE+EXCLUSION | Search MINORITY+CYTOTYPE+EXCLUSION |
| 7 | CYTOGEOGRAPHY | 9 | 38% | 1% | 20 | Search CYTOGEOGRAPHY | Search CYTOGEOGRAPHY |
| 8 | EXPRESSION LEVEL DOMINANCE | 8 | 100% | 0% | 5 | Search EXPRESSION+LEVEL+DOMINANCE | Search EXPRESSION+LEVEL+DOMINANCE |
| 9 | ALLOPOLYPLOID | 8 | 24% | 2% | 28 | Search ALLOPOLYPLOID | Search ALLOPOLYPLOID |
| 10 | GENOME DOUBLING | 7 | 53% | 1% | 9 | Search GENOME+DOUBLING | Search GENOME+DOUBLING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ARABIDOPSIS ALLOTETRAPLOIDS | 104 | 75% | 6% | 76 |
| 2 | HABITAT DIFFERENTIATION | 98 | 84% | 4% | 53 |
| 3 | RESYNTHESIZED BRASSICA NAPUS | 96 | 88% | 3% | 45 |
| 4 | BUTTERCUPS RANUNCULUS ADONEUS | 76 | 100% | 2% | 24 |
| 5 | HEXAPLOID CYTOTYPES | 55 | 87% | 2% | 27 |
| 6 | POLYPLOIDY | 53 | 21% | 17% | 229 |
| 7 | TRAGOPOGON MISCELLUS ASTERACEAE | 49 | 82% | 2% | 28 |
| 8 | AUTOPOLYPLOID PLANTAGO MEDIA | 45 | 94% | 1% | 16 |
| 9 | CHAMERION ANGUSTIFOLIUM ONAGRACEAE | 44 | 85% | 2% | 23 |
| 10 | NEWLY SYNTHESIZED AMPHIPLOIDS | 39 | 53% | 4% | 52 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Polyploidy and its effect on evolutionary success: old questions revisited with new tools | 2013 | 46 | 60 | 82% |
| Polyploidy: adaptation to the genomic environment | 2015 | 2 | 29 | 48% |
| Unreduced gametes: meiotic mishap or evolutionary mechanism? | 2015 | 3 | 60 | 30% |
| Evolutionary consequences of autopolyploidy | 2010 | 148 | 97 | 79% |
| THE POLYPLOIDY REVOLUTION THEN...AND NOW: STEBBINS REVISITED | 2014 | 17 | 198 | 62% |
| The Role of Hybridization in Plant Speciation | 2009 | 296 | 135 | 44% |
| The more the better? The role of polyploidy in facilitating plant invasions | 2012 | 70 | 270 | 41% |
| Ecological studies of polyploidy in the 100 years following its discovery | 2014 | 8 | 152 | 73% |
| Neopolyploidy in flowering plants | 2002 | 375 | 51 | 63% |
| Recent progress and challenges in population genetics of polyploid organisms: an overview of current state-of-the-art molecular and statistical tools | 2014 | 21 | 209 | 31% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PLANT BIOL GENOME | 4 | 67% | 0.3% | 4 |
| 2 | PLANT BIOSYSTEMAT ECOL RG | 4 | 67% | 0.3% | 4 |
| 3 | PLANTENGENET VEREDELING | 4 | 75% | 0.2% | 3 |
| 4 | MOL SYSTEMAT EVOLUTIONARY GENET | 3 | 100% | 0.2% | 3 |
| 5 | MOL EPIGENET | 3 | 12% | 1.6% | 22 |
| 6 | GLORIA COORDINAT | 2 | 67% | 0.1% | 2 |
| 7 | GRP POPULAT SPECIES EVOLUT | 2 | 67% | 0.1% | 2 |
| 8 | INTER EGIATE GENET PROGRAM | 2 | 67% | 0.1% | 2 |
| 9 | GENOME EVOLUT SPECIAT | 1 | 50% | 0.1% | 2 |
| 10 | PLANT BIOSYST ECOL RG | 1 | 100% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000179982 | CHROMOSOME DOUBLING//TETRAPLOID//ENDOSPERM CULTURE |
| 2 | 0.0000158137 | ARTEMISIINAE//ANTHEMIDEAE//TRIPLEUROSPERMUM |
| 3 | 0.0000152694 | PLANT MICROBIAL GENOM//LRT1//NONADDITIVE PROTEIN |
| 4 | 0.0000140082 | GENOME SIZE//C VALUE//NUCLEAR DNA CONTENT |
| 5 | 0.0000134570 | DRABA//BRASSICACEAE//S SIBERIAN BOT GARDEN |
| 6 | 0.0000133856 | WHOLE GENOME DUPLICATION//GENET MOL CELLULAIRE ECT LEVU INTERE//GENOME DUPLICATION |
| 7 | 0.0000132729 | CHRYSOSPLENIUM//LASTHENIA CALIFORNICA//SAXIFRAGACEAE |
| 8 | 0.0000128087 | MICROSERIS//GROUNDSEL SENECIO VULGARIS//MICROSERIS DOUGLASII |
| 9 | 0.0000117423 | HYBRID LETHALITY//HYBRID NECROSIS//NICOTIANA SECTION SUAVEOLENTES |
| 10 | 0.0000116505 | RDNA INTERGENIC SPACER//NON CONCERTED EVOLUTION//INTERGENIC SPACER |