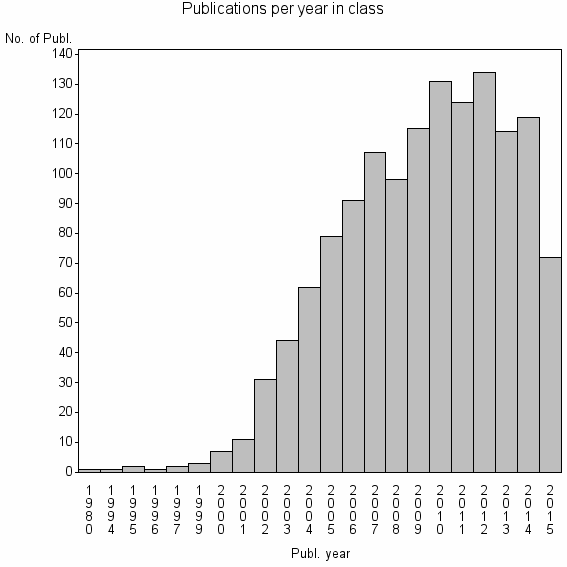
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 6944 | 1349 | 39.2 | 70% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 584 | 13517 | BMC SYSTEMS BIOLOGY//FLUX BALANCE ANALYSIS//SYNTHETIC BIOLOGY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | NETWORK INFERENCE | Author keyword | 16 | 34% | 3% | 38 |
| 2 | GENE REGULATORY NETWORKS | Author keyword | 13 | 16% | 5% | 71 |
| 3 | GENE NETWORK INFERENCE | Author keyword | 8 | 75% | 0% | 6 |
| 4 | GENE REGULATORY NETWORK | Author keyword | 7 | 12% | 4% | 53 |
| 5 | DREAM CHALLENGE | Author keyword | 6 | 100% | 0% | 4 |
| 6 | GENE ASSOCIATION NETWORKS | Author keyword | 4 | 75% | 0% | 3 |
| 7 | NESTED EFFECTS MODELS | Author keyword | 4 | 75% | 0% | 3 |
| 8 | COMPUTAT BIOL MACHINE LEARNING | Address | 4 | 28% | 1% | 11 |
| 9 | NETGENERATOR | Author keyword | 3 | 100% | 0% | 3 |
| 10 | SYST BIOL BIOCHEM MOL BIOPHYS | Address | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NETWORK INFERENCE | 16 | 34% | 3% | 38 | Search NETWORK+INFERENCE | Search NETWORK+INFERENCE |
| 2 | GENE REGULATORY NETWORKS | 13 | 16% | 5% | 71 | Search GENE+REGULATORY+NETWORKS | Search GENE+REGULATORY+NETWORKS |
| 3 | GENE NETWORK INFERENCE | 8 | 75% | 0% | 6 | Search GENE+NETWORK+INFERENCE | Search GENE+NETWORK+INFERENCE |
| 4 | GENE REGULATORY NETWORK | 7 | 12% | 4% | 53 | Search GENE+REGULATORY+NETWORK | Search GENE+REGULATORY+NETWORK |
| 5 | DREAM CHALLENGE | 6 | 100% | 0% | 4 | Search DREAM+CHALLENGE | Search DREAM+CHALLENGE |
| 6 | GENE ASSOCIATION NETWORKS | 4 | 75% | 0% | 3 | Search GENE+ASSOCIATION+NETWORKS | Search GENE+ASSOCIATION+NETWORKS |
| 7 | NESTED EFFECTS MODELS | 4 | 75% | 0% | 3 | Search NESTED+EFFECTS+MODELS | Search NESTED+EFFECTS+MODELS |
| 8 | NETGENERATOR | 3 | 100% | 0% | 3 | Search NETGENERATOR | Search NETGENERATOR |
| 9 | SET MULTICOVER | 3 | 60% | 0% | 3 | Search SET+MULTICOVER | Search SET+MULTICOVER |
| 10 | TIME COURSE GENE EXPRESSION DATA | 2 | 50% | 0% | 3 | Search TIME+COURSE+GENE+EXPRESSION+DATA | Search TIME+COURSE+GENE+EXPRESSION+DATA |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | COMPOUND MODE | 42 | 66% | 3% | 39 |
| 2 | NESTED EFFECTS MODELS | 33 | 100% | 1% | 13 |
| 3 | EXPRESSION DATA | 32 | 13% | 17% | 224 |
| 4 | REGULATORY NETWORKS | 28 | 12% | 16% | 219 |
| 5 | BAYESIAN NETWORKS | 21 | 16% | 9% | 127 |
| 6 | GENE REGULATORY NETWORKS | 17 | 18% | 6% | 84 |
| 7 | MODULE NETWORKS | 14 | 53% | 1% | 18 |
| 8 | RELEVANCE NETWORKS | 10 | 73% | 1% | 8 |
| 9 | ARACNE | 9 | 64% | 1% | 9 |
| 10 | ENGINEERING GENE NETWORKS | 7 | 43% | 1% | 13 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Gene regulatory network inference: Data integration in dynamic models-A review | 2009 | 181 | 73 | 59% |
| How to infer gene networks from expression profiles | 2007 | 282 | 22 | 59% |
| Inferring cellular networks using probabilistic graphical models | 2004 | 474 | 15 | 33% |
| How to infer gene networks from expression profiles, revisited | 2011 | 26 | 34 | 59% |
| Advantages and limitations of current network inference methods | 2010 | 105 | 103 | 32% |
| Computational methods for discovering gene networks from expression data | 2009 | 55 | 83 | 47% |
| Inference of Gene Regulatory Networks Using Time-Series Data: A Survey | 2009 | 27 | 71 | 61% |
| Modelling and analysis of gene regulatory networks | 2008 | 205 | 102 | 13% |
| Reverse engineering and verification of gene networks: Principles, assumptions, and limitations of present methods and future perspectives | 2009 | 32 | 111 | 47% |
| Biological network inference for drug discovery | 2013 | 7 | 32 | 31% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | COMPUTAT BIOL MACHINE LEARNING | 4 | 28% | 0.8% | 11 |
| 2 | SYST BIOL BIOCHEM MOL BIOPHYS | 3 | 100% | 0.2% | 3 |
| 3 | COMPUTAT ASTROBIOL FUNDAMENTAL BIOL | 2 | 67% | 0.1% | 2 |
| 4 | INFECT DIS MOL MEDMED | 2 | 67% | 0.1% | 2 |
| 5 | JBI | 2 | 67% | 0.1% | 2 |
| 6 | KNOWLEDGE DISCOVERY BIOINFORMAT GRP | 2 | 50% | 0.2% | 3 |
| 7 | JOINT SYST BIOL | 2 | 18% | 0.6% | 8 |
| 8 | SCI COMP COMPLEX SYST MODELLING | 1 | 38% | 0.2% | 3 |
| 9 | ADM INFORMAT SYST MANAGEMENT | 1 | 50% | 0.1% | 2 |
| 10 | GENET GAETANO SALVATORE | 1 | 50% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000224634 | BICLUSTERING//GENE EXPRESSION DATA//GENE EXPRESSION DATA ANALYSIS |
| 2 | 0.0000186813 | BOOLEAN NETWORKS//SEMI TENSOR PRODUCT//BOOLEAN NETWORK |
| 3 | 0.0000163257 | PROGRAM COMPUTAT GENOM//CIFN//PROGRAMA GENOM COMPUTAC |
| 4 | 0.0000162095 | BIOCHEMICAL SYSTEMS THEORY//S SYSTEM//S DISTRIBUTION |
| 5 | 0.0000137653 | GENETIC REGULATORY NETWORKS GRNS//GENETIC REGULATORY NETWORKS//GENETIC REGULATORY NETWORK |
| 6 | 0.0000100055 | EDGE BIOMARKER//SIBS NOVO NORDISK TRANSLAT PREDIABET//RESP18 |
| 7 | 0.0000097095 | GENE SET ANALYSIS//ENRICHMENT ANALYSIS//FUNCT GENOM NODE |
| 8 | 0.0000095897 | MOTIF DISCOVERY//CLOSEST STRING PROBLEM//MOTIF FINDING |
| 9 | 0.0000095796 | CIS MOTIF//ARABIDOPSIS T87 CELL LINE//GENE ATLAS |
| 10 | 0.0000095499 | PROTEIN INTERACTION NETWORKS//PROTEIN PROTEIN INTERACTION NETWORKS//PROTEIN PROTEIN INTERACTION NETWORK |