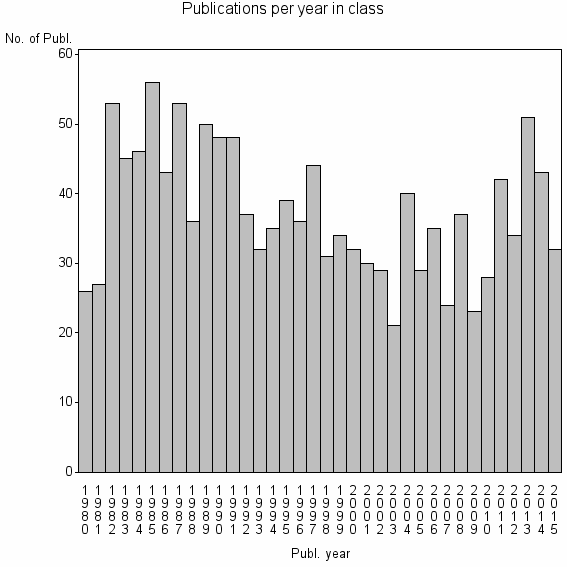
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 6948 | 1349 | 41.8 | 76% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1031 | 9720 | NUCLEOLUS//RIBOSOME BIOGENESIS//NUCLEOLAR ORGANIZER REGIONS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MOL BIOL CELL 2 | Address | 68 | 69% | 4% | 58 |
| 2 | RNA POLYMERASE I | Author keyword | 67 | 48% | 7% | 101 |
| 3 | UBF | Author keyword | 22 | 50% | 2% | 32 |
| 4 | UPSTREAM BINDING FACTOR | Author keyword | 17 | 72% | 1% | 13 |
| 5 | SL1 | Author keyword | 12 | 75% | 1% | 9 |
| 6 | RRN3 | Author keyword | 8 | 100% | 0% | 5 |
| 7 | NORC | Author keyword | 7 | 53% | 1% | 10 |
| 8 | TIF IA | Author keyword | 6 | 80% | 0% | 4 |
| 9 | TIP5 | Author keyword | 6 | 71% | 0% | 5 |
| 10 | RDNA TRANSCRIPTION | Author keyword | 6 | 36% | 1% | 14 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA POLYMERASE I | 67 | 48% | 7% | 101 | Search RNA+POLYMERASE+I | Search RNA+POLYMERASE+I |
| 2 | UBF | 22 | 50% | 2% | 32 | Search UBF | Search UBF |
| 3 | UPSTREAM BINDING FACTOR | 17 | 72% | 1% | 13 | Search UPSTREAM+BINDING+FACTOR | Search UPSTREAM+BINDING+FACTOR |
| 4 | SL1 | 12 | 75% | 1% | 9 | Search SL1 | Search SL1 |
| 5 | RRN3 | 8 | 100% | 0% | 5 | Search RRN3 | Search RRN3 |
| 6 | NORC | 7 | 53% | 1% | 10 | Search NORC | Search NORC |
| 7 | TIF IA | 6 | 80% | 0% | 4 | Search TIF+IA | Search TIF+IA |
| 8 | TIP5 | 6 | 71% | 0% | 5 | Search TIP5 | Search TIP5 |
| 9 | RDNA TRANSCRIPTION | 6 | 36% | 1% | 14 | Search RDNA+TRANSCRIPTION | Search RDNA+TRANSCRIPTION |
| 10 | CX 5461 | 4 | 75% | 0% | 3 | Search CX+5461 | Search CX+5461 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FACTOR UBF | 114 | 73% | 6% | 87 |
| 2 | UPSTREAM ACTIVATION FACTOR | 81 | 96% | 2% | 25 |
| 3 | FACTOR TIF IA | 76 | 69% | 5% | 65 |
| 4 | RDNA TRANSCRIPTION | 73 | 52% | 7% | 100 |
| 5 | MOUSE RDNA PROMOTER | 65 | 95% | 2% | 21 |
| 6 | RNA POLYMERASE I | 61 | 24% | 16% | 220 |
| 7 | FACTOR XUBF | 49 | 94% | 1% | 17 |
| 8 | MOUSE RDNA | 38 | 86% | 1% | 19 |
| 9 | RIBOSOMAL GENE TRANSCRIPTION | 34 | 55% | 3% | 43 |
| 10 | POLYMERASE I TRANSCRIPTION | 31 | 28% | 7% | 95 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The Epigenetics of rRNA Genes: From Molecular to Chromosome Biology | 2008 | 173 | 129 | 60% |
| Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus | 2003 | 264 | 91 | 69% |
| The RNA Polymerase I Transcription Machinery: An Emerging Target for the Treatment of Cancer | 2010 | 123 | 122 | 52% |
| Regulation of mammalian ribosomal gene transcription by RNA polymerase I | 1999 | 155 | 207 | 68% |
| A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis | 2007 | 99 | 217 | 65% |
| Regulation of rDNA transcription in response to growth factors, nutrients and energy | 2015 | 1 | 104 | 34% |
| At the center of eukaryotic life | 2002 | 107 | 17 | 71% |
| RNA-polymerase-I-directed rDNA transcription, life and works | 2005 | 113 | 82 | 67% |
| Epigenetic control of RNA polymerase I transcription in mammalian cells | 2013 | 18 | 102 | 50% |
| PROMOTION AND REGULATION OF RIBOSOMAL TRANSCRIPTION IN EUKARYOTES BY RNA-POLYMERASE-I | 1995 | 160 | 265 | 66% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOL BIOL CELL 2 | 68 | 69% | 4.3% | 58 |
| 2 | HENRY HOOD PROGRAM | 3 | 27% | 0.7% | 9 |
| 3 | UNITE REGULAT TRANSCRIPT MALAD GENET | 3 | 60% | 0.2% | 3 |
| 4 | UNITE PROPRE RECH 2228 | 2 | 44% | 0.3% | 4 |
| 5 | LEHRSTUHL BIOCHEM 3 | 2 | 32% | 0.4% | 6 |
| 6 | INTERDISCIPLINAIRE SCI VIVANT ST PE | 1 | 38% | 0.2% | 3 |
| 7 | MOL BIOL CELL II | 1 | 100% | 0.1% | 2 |
| 8 | WORLD CLASS UNIV NEUROCYT GRP | 1 | 100% | 0.1% | 2 |
| 9 | BIOCHEM GENET MIKROBIOL | 1 | 18% | 0.4% | 6 |
| 10 | KY SPINAL CORD INJURY | 1 | 40% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000224915 | RDNA INTERGENIC SPACER//NON CONCERTED EVOLUTION//INTERGENIC SPACER |
| 2 | 0.0000213755 | POKEY//LAMPBRUSH LOOPS//FERTILITY GENES |
| 3 | 0.0000109208 | SNORNA//SMALL NUCLEOLAR RNAS//PRE RRNA PROCESSING |
| 4 | 0.0000100493 | CHROMATIN DIS//RIBOSOMAL RNA SYNTHESIS//CISPLATIN RNA ADDUCTS |
| 5 | 0.0000090142 | NUCLEOLUS//COILIN//NUCLEOLOGENESIS |
| 6 | 0.0000088559 | RNA POLYMERASE III//TFIIIB//SNAPC |
| 7 | 0.0000086132 | HETEROCHROMATINIZATION//SATELLITE ASSOCIATION//ECTOPIC NUCLEOLUS ORGANIZER REGION |
| 8 | 0.0000082025 | NUCLEOLIN//SURFACE NUCLEOLIN//TIP ALPHA |
| 9 | 0.0000080217 | RPB7//RPB4//INTERSPECIFIC COMPLEMENTATION |
| 10 | 0.0000064405 | SIR COMPLEX//SIR3//SIR PROTEINS |