Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
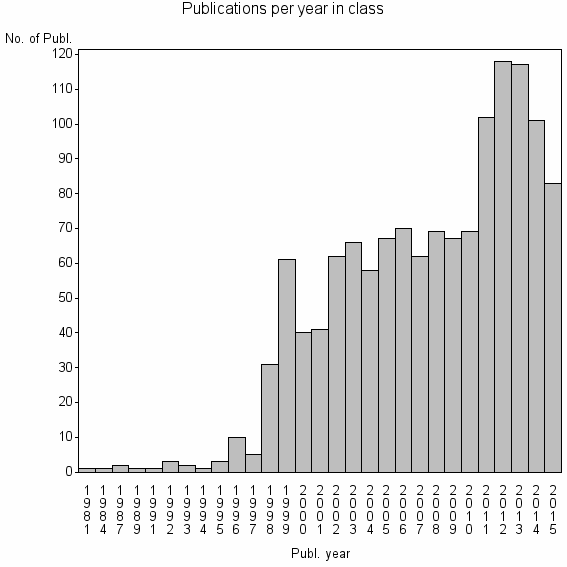
| 7245 | 1314 | 48.1 | 94% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | COP9 SIGNALOSOME | Author keyword | 103 | 78% | 5% | 69 |
| 2 | NEDD8 | Author keyword | 96 | 66% | 7% | 90 |
| 3 | NEDDYLATION | Author keyword | 51 | 69% | 3% | 43 |
| 4 | CULLIN | Author keyword | 47 | 57% | 4% | 55 |
| 5 | MLN4924 | Author keyword | 28 | 69% | 2% | 24 |
| 6 | CSN6 | Author keyword | 23 | 100% | 1% | 10 |
| 7 | DENEDDYLATION | Author keyword | 23 | 100% | 1% | 10 |
| 8 | 3 M SYNDROME | Author keyword | 21 | 90% | 1% | 9 |
| 9 | CULLINS | Author keyword | 17 | 79% | 1% | 11 |
| 10 | CSN5 | Author keyword | 16 | 65% | 1% | 15 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | COP9 SIGNALOSOME | 103 | 78% | 5% | 69 | Search COP9+SIGNALOSOME | Search COP9+SIGNALOSOME |
| 2 | NEDD8 | 96 | 66% | 7% | 90 | Search NEDD8 | Search NEDD8 |
| 3 | NEDDYLATION | 51 | 69% | 3% | 43 | Search NEDDYLATION | Search NEDDYLATION |
| 4 | CULLIN | 47 | 57% | 4% | 55 | Search CULLIN | Search CULLIN |
| 5 | MLN4924 | 28 | 69% | 2% | 24 | Search MLN4924 | Search MLN4924 |
| 6 | CSN6 | 23 | 100% | 1% | 10 | Search CSN6 | Search CSN6 |
| 7 | DENEDDYLATION | 23 | 100% | 1% | 10 | Search DENEDDYLATION | Search DENEDDYLATION |
| 8 | 3 M SYNDROME | 21 | 90% | 1% | 9 | Search 3+M+SYNDROME | Search 3+M+SYNDROME |
| 9 | CULLINS | 17 | 79% | 1% | 11 | Search CULLINS | Search CULLINS |
| 10 | CSN5 | 16 | 65% | 1% | 15 | Search CSN5 | Search CSN5 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NEDD8 | 95 | 58% | 8% | 109 |
| 2 | COP9 SIGNALOSOME | 84 | 35% | 15% | 194 |
| 3 | CUL1 | 59 | 77% | 3% | 40 |
| 4 | NEDD8 MODIFICATION | 57 | 79% | 3% | 37 |
| 5 | CULLIN | 52 | 80% | 2% | 32 |
| 6 | NEDDYLATION | 50 | 81% | 2% | 30 |
| 7 | NEDD8 CONJUGATION | 46 | 75% | 3% | 33 |
| 8 | CUL7 | 42 | 83% | 2% | 24 |
| 9 | NEDD8 ACTIVATING ENZYME INHIBITOR | 42 | 78% | 2% | 28 |
| 10 | NEDD8 CONJUGATION PATHWAY | 38 | 89% | 1% | 17 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Protein neddylation: beyond cullin-RING ligases | 2015 | 7 | 176 | 61% |
| Function and regulation of Cullin-RING ubiquitin ligases | 2005 | 922 | 137 | 48% |
| The cullin protein family | 2011 | 89 | 94 | 60% |
| Building and remodelling Cullin-RING E3 ubiquitin ligases - 'Ubiquitylation: mechanism and functions' review series | 2013 | 25 | 116 | 60% |
| The COP9 signalosome: more than a protease | 2008 | 163 | 85 | 78% |
| Regulation of cancer-related pathways by protein NEDDylation and strategies for the use of NEDD8 inhibitors in the clinic | 2015 | 1 | 173 | 65% |
| Function and regulation of protein neddylation - 'Protein Modifications: Beyond the Usual Suspects' review series | 2008 | 118 | 94 | 79% |
| NEDD8 Pathways in Cancer, Sine Quibus Non | 2011 | 45 | 113 | 61% |
| Cullin-RING ubiquitin ligases: global regulation and activation cycles | 2008 | 131 | 121 | 73% |
| The Cullin-RING Ubiquitin-Protein Ligases | 2011 | 95 | 231 | 30% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PEALE SCI | 9 | 83% | 0.4% | 5 |
| 2 | PROT UBIQUITYLAT UNIT | 6 | 100% | 0.3% | 4 |
| 3 | EXPT TERATOLMINIST EDUC | 3 | 30% | 0.6% | 8 |
| 4 | ROBERT HOLLY | 3 | 60% | 0.2% | 3 |
| 5 | ACUTE LUNG INJURY EXCELLENCE | 3 | 24% | 0.8% | 10 |
| 6 | SCOTTISH CELL SIGNALLING | 3 | 35% | 0.5% | 6 |
| 7 | ABT ENTWICKLUNGSGENET | 2 | 67% | 0.2% | 2 |
| 8 | BEIJING GENE ENGN DRUG BIOTECHNOL | 2 | 67% | 0.2% | 2 |
| 9 | HUMAN GENET HUMAN GENOME | 2 | 67% | 0.2% | 2 |
| 10 | MOL CH ERONE BIOL | 2 | 67% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000288094 | FBXW7//FBW7//HCDC4 |
| 2 | 0.0000194078 | U BOX//CELLULAR BIOL MOL GENET//PLANT BIOL GRP PROGRAM |
| 3 | 0.0000180853 | DSK2//UBIQUILIN 1//LUBAC |
| 4 | 0.0000129383 | SKP2//P27KIP1//P27 |
| 5 | 0.0000101601 | TNFAIP1//KCTD7//KCTD10 |
| 6 | 0.0000085806 | PUPYLATION//PROKARYOTIC UBIQUITIN LIKE PROTEIN//URM1 |
| 7 | 0.0000077360 | CXCL14//DBC2//BRAK |
| 8 | 0.0000072433 | UBCH10//ANAPHASE PROMOTING COMPLEX//GREATWALL |
| 9 | 0.0000066626 | USP22//UCH L1//DEUBIQUITINATING ENZYME |
| 10 | 0.0000060897 | POLYUBIQUITIN GENE//UBIQUITYL CALMODULIN SYNTHETASE//FREE UBIQUITIN |