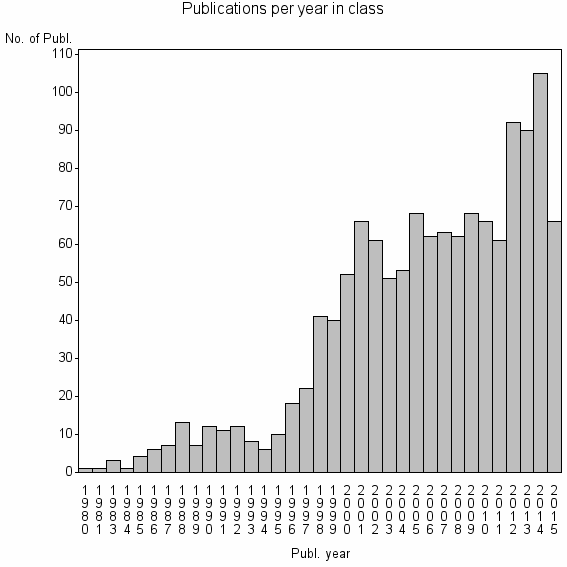
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 7283 | 1309 | 55.8 | 92% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PIKFYVE | Author keyword | 52 | 80% | 2% | 32 |
| 2 | PTDINS5P | Author keyword | 27 | 92% | 1% | 11 |
| 3 | GOLPH3 | Author keyword | 23 | 76% | 1% | 16 |
| 4 | YUNIS VARON SYNDROME | Author keyword | 21 | 90% | 1% | 9 |
| 5 | PHOSPHOINOSITIDE | Author keyword | 21 | 19% | 8% | 100 |
| 6 | SECT MOL SIGNAL TRANSDUCT | Address | 16 | 48% | 2% | 24 |
| 7 | FAB1 | Author keyword | 15 | 88% | 1% | 7 |
| 8 | PI 4 KINASE | Author keyword | 13 | 69% | 1% | 11 |
| 9 | PHOSPHATIDYLINOSITOL 4 KINASE | Author keyword | 12 | 43% | 2% | 21 |
| 10 | FIG4 | Author keyword | 11 | 69% | 1% | 9 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PIKFYVE | 52 | 80% | 2% | 32 | Search PIKFYVE | Search PIKFYVE |
| 2 | PTDINS5P | 27 | 92% | 1% | 11 | Search PTDINS5P | Search PTDINS5P |
| 3 | GOLPH3 | 23 | 76% | 1% | 16 | Search GOLPH3 | Search GOLPH3 |
| 4 | YUNIS VARON SYNDROME | 21 | 90% | 1% | 9 | Search YUNIS+VARON+SYNDROME | Search YUNIS+VARON+SYNDROME |
| 5 | PHOSPHOINOSITIDE | 21 | 19% | 8% | 100 | Search PHOSPHOINOSITIDE | Search PHOSPHOINOSITIDE |
| 6 | FAB1 | 15 | 88% | 1% | 7 | Search FAB1 | Search FAB1 |
| 7 | PI 4 KINASE | 13 | 69% | 1% | 11 | Search PI+4+KINASE | Search PI+4+KINASE |
| 8 | PHOSPHATIDYLINOSITOL 4 KINASE | 12 | 43% | 2% | 21 | Search PHOSPHATIDYLINOSITOL+4+KINASE | Search PHOSPHATIDYLINOSITOL+4+KINASE |
| 9 | FIG4 | 11 | 69% | 1% | 9 | Search FIG4 | Search FIG4 |
| 10 | PTDINS3 5P 2 | 11 | 78% | 1% | 7 | Search PTDINS3+5P+2 | Search PTDINS3+5P+2 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MEMBRANE HOMEOSTASIS | 68 | 84% | 3% | 37 |
| 2 | 4 PHOSPHATE 5 KINASE | 45 | 63% | 3% | 45 |
| 3 | PHOSPHATIDYLINOSITOL 3 5 BISPHOSPHATE | 43 | 60% | 4% | 47 |
| 4 | PHOSPHATE KINASE | 41 | 65% | 3% | 40 |
| 5 | PIKFYVE | 39 | 68% | 3% | 34 |
| 6 | 4 KINASE | 30 | 57% | 3% | 35 |
| 7 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE | 30 | 26% | 7% | 97 |
| 8 | LIPID KINASE FAMILY | 29 | 66% | 2% | 27 |
| 9 | I GAMMA | 27 | 83% | 1% | 15 |
| 10 | FAB1P | 27 | 60% | 2% | 29 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Phosphoinositides in cell regulation and membrane dynamics | 2006 | 984 | 70 | 47% |
| Membrane recognition by phospholipid-binding domains | 2008 | 588 | 114 | 27% |
| Polyphosphoinositide binding domains: Key to inositol lipid biology | 2015 | 2 | 155 | 47% |
| PHOSPHOINOSITIDES: TINY LIPIDS WITH GIANT IMPACT ON CELL REGULATION | 2013 | 103 | 1708 | 19% |
| GOLPH3 Links the Golgi, DNA Damage, and Cancer | 2015 | 1 | 39 | 67% |
| Phosphatidylinositol-3,5-Bisphosphate: No Longer the Poor PIP2 | 2012 | 27 | 80 | 79% |
| Phosphoinositides regulate ion channels | 2015 | 6 | 132 | 13% |
| Myotubularin phosphoinositide phosphatases: cellular functions and disease pathophysiology | 2012 | 34 | 79 | 58% |
| Coordination of Golgi functions by phosphatidylinositol 4-kinases | 2011 | 63 | 90 | 36% |
| Translation of the phosphoinositide code by PI effectors | 2010 | 72 | 96 | 44% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SECT MOL SIGNAL TRANSDUCT | 16 | 48% | 1.8% | 24 |
| 2 | CRUK INOSITIDE | 9 | 83% | 0.4% | 5 |
| 3 | PLASMA MEMBRANE RECEPTORS | 3 | 60% | 0.2% | 3 |
| 4 | MOL DYNAM SYN T FUNCT | 2 | 38% | 0.4% | 5 |
| 5 | CANC UK INOSITIDE | 2 | 67% | 0.2% | 2 |
| 6 | PROGRAM DEV NEUROSCI | 2 | 12% | 1.2% | 16 |
| 7 | WADHWANI | 2 | 50% | 0.2% | 3 |
| 8 | INOSITIDE | 2 | 12% | 1.1% | 14 |
| 9 | HUMAN INHERITED NEUROPATHIES UNIT | 1 | 100% | 0.2% | 2 |
| 10 | LIPID MEMBRANE BIOL GRP | 1 | 100% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000204195 | BIOL CHEMBUNKYO KU//DEMETHOXYVIRIDIN//2 4 MORPHOLINYL 8 PHENYLCHROMONE |
| 2 | 0.0000186225 | LOWE SYNDROME//OCULOCEREBRORENAL SYNDROME//OCRL1 |
| 3 | 0.0000155714 | OXYSTEROL BINDING PROTEIN//OXYSTEROL BINDING PROTEINS//OSBP |
| 4 | 0.0000154341 | RETROMER//SORTING NEXIN//VPS35 |
| 5 | 0.0000141057 | SHIP2//INOSITOL 5 PHOSPHATASE//SHIP1 |
| 6 | 0.0000116250 | SECT PLANT PHYSIOL//PHOSPHATIDIC ACID//PHOSPHOLIPID SIGNALLING |
| 7 | 0.0000098887 | STEROL CARRIER PROTEIN 2//PHOSPHATIDYLINOSITOL TRANSFER PROTEIN//GLYCOLIPID TRANSFER PROTEIN |
| 8 | 0.0000089739 | DNA BOUND LIPIDS//NUCLEAR PHOSPHOLIPIDS//PIKE A |
| 9 | 0.0000085316 | ADP RIBOSYLATION FACTOR//ARF6//CYTOHESIN |
| 10 | 0.0000084065 | PHOSPHOLIPASE C DELTA 1//PHOSPHOLIPASE C GAMMA 1//GENOME BIOSIGNAL |