Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
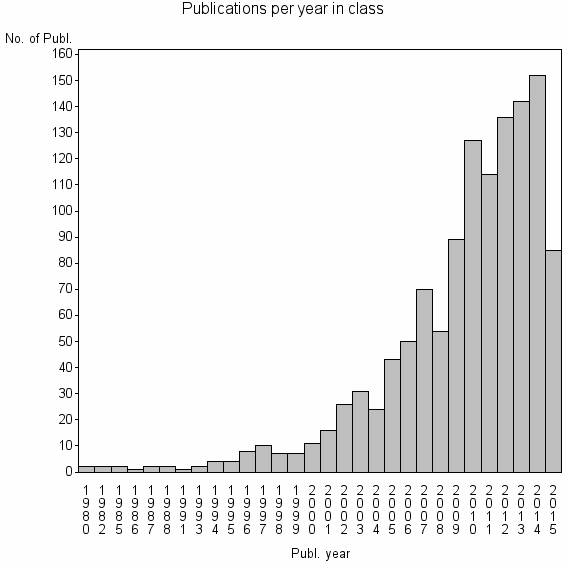
| 8044 | 1224 | 54.9 | 91% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | HFQ | Author keyword | 191 | 79% | 10% | 123 |
| 2 | S RNA | Author keyword | 46 | 52% | 5% | 64 |
| 3 | RYHB | Author keyword | 29 | 75% | 2% | 21 |
| 4 | RIBOREGULATION | Author keyword | 23 | 76% | 1% | 16 |
| 5 | RNA BIOL GRP | Address | 19 | 47% | 2% | 30 |
| 6 | DSRA | Author keyword | 18 | 65% | 1% | 17 |
| 7 | SRNAS | Author keyword | 17 | 75% | 1% | 12 |
| 8 | HFQ PROTEIN | Author keyword | 15 | 88% | 1% | 7 |
| 9 | CSRA | Author keyword | 14 | 65% | 1% | 13 |
| 10 | CSRB | Author keyword | 12 | 86% | 0% | 6 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HFQ | 191 | 79% | 10% | 123 | Search HFQ | Search HFQ |
| 2 | S RNA | 46 | 52% | 5% | 64 | Search S+RNA | Search S+RNA |
| 3 | RYHB | 29 | 75% | 2% | 21 | Search RYHB | Search RYHB |
| 4 | RIBOREGULATION | 23 | 76% | 1% | 16 | Search RIBOREGULATION | Search RIBOREGULATION |
| 5 | DSRA | 18 | 65% | 1% | 17 | Search DSRA | Search DSRA |
| 6 | SRNAS | 17 | 75% | 1% | 12 | Search SRNAS | Search SRNAS |
| 7 | HFQ PROTEIN | 15 | 88% | 1% | 7 | Search HFQ+PROTEIN | Search HFQ+PROTEIN |
| 8 | CSRA | 14 | 65% | 1% | 13 | Search CSRA | Search CSRA |
| 9 | CSRB | 12 | 86% | 0% | 6 | Search CSRB | Search CSRB |
| 10 | OXYS | 12 | 86% | 0% | 6 | Search OXYS | Search OXYS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SM LIKE PROTEIN | 311 | 84% | 14% | 171 |
| 2 | ESCHERICHIA COLI HFQ | 222 | 91% | 8% | 92 |
| 3 | SMALL NONCODING RNAS | 174 | 61% | 15% | 187 |
| 4 | HFQ | 135 | 69% | 9% | 116 |
| 5 | SMALL REGULATORY RNA | 134 | 83% | 6% | 75 |
| 6 | CHAPERONE HFQ | 132 | 94% | 4% | 47 |
| 7 | SOLUBLE RNAS | 102 | 89% | 4% | 47 |
| 8 | BACTERIAL SMALL RNA | 87 | 88% | 3% | 42 |
| 9 | BINDING PROTEIN HFQ | 77 | 89% | 3% | 34 |
| 10 | SOLUBLE RNA | 65 | 68% | 5% | 57 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Regulation by Small RNAs in Bacteria: Expanding Frontiers | 2011 | 250 | 106 | 83% |
| Hfq and its constellation of RNA | 2011 | 239 | 146 | 79% |
| Regulatory small RNAs from the 3 ' regions of bacterial mRNAs | 2015 | 4 | 57 | 79% |
| Regulatory RNAs in Bacteria | 2009 | 566 | 116 | 64% |
| Bacterial Small RNA-based Negative Regulation: Hfq and Its Accomplices | 2013 | 52 | 56 | 77% |
| Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems | 2013 | 64 | 93 | 46% |
| Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA | 2015 | 2 | 88 | 68% |
| The role of Hfq in bacterial pathogens | 2010 | 124 | 63 | 78% |
| Hfq structure, function and ligand binding | 2007 | 207 | 51 | 84% |
| Regulatory RNAS and target mRNA decay in prokaryotes | 2013 | 26 | 71 | 75% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RNA BIOL GRP | 19 | 47% | 2.5% | 30 |
| 2 | PROGRAM CELLULAR MOL DEV BIOL BIOPHYS | 6 | 100% | 0.3% | 4 |
| 3 | MOL INFECT BIOL | 4 | 11% | 2.9% | 36 |
| 4 | CNRS FRE3630 | 4 | 75% | 0.2% | 3 |
| 5 | PARKER H PETIT BIOSCI BIOENGN | 4 | 75% | 0.2% | 3 |
| 6 | PROGRAMA INTERACC BIOL | 4 | 56% | 0.4% | 5 |
| 7 | UP EA2311 | 4 | 56% | 0.4% | 5 |
| 8 | AG BAKTERIENGENET | 4 | 31% | 0.8% | 10 |
| 9 | GRP ECOL GENET RIZOSFERA | 4 | 46% | 0.5% | 6 |
| 10 | FRE 3132 | 3 | 100% | 0.2% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000173038 | RNASE E//RNASE G//POLYNUCLEOTIDE PHOSPHORYLASE |
| 2 | 0.0000108917 | RIBOSWITCH//RIBOSWITCHES//STREPTOMYCES DAVAWENSIS |
| 3 | 0.0000105377 | PPGPP//STRINGENT RESPONSE//RPOS |
| 4 | 0.0000087675 | ENVZ//OMPR//ENVZ OSMOSENSOR |
| 5 | 0.0000076072 | PROGRAM COMPUTAT GENOM//CIFN//PROGRAMA GENOM COMPUTAC |
| 6 | 0.0000069429 | RNA SECONDARY STRUCTURE//RNA SECONDARY STRUCTURE PREDICTION//RNA STRUCTURE PREDICTION |
| 7 | 0.0000068199 | TOXIN ANTITOXIN//TOXIN ANTITOXIN SYSTEM//TOXIN ANTITOXIN SYSTEMS |
| 8 | 0.0000065879 | FERRIC UPTAKE REGULATOR//DTXR//FUR BOX |
| 9 | 0.0000063419 | TRANS TRANSLATION//TMRNA//SMPB |
| 10 | 0.0000060759 | HTRA1//HTRA//HTRA2 |