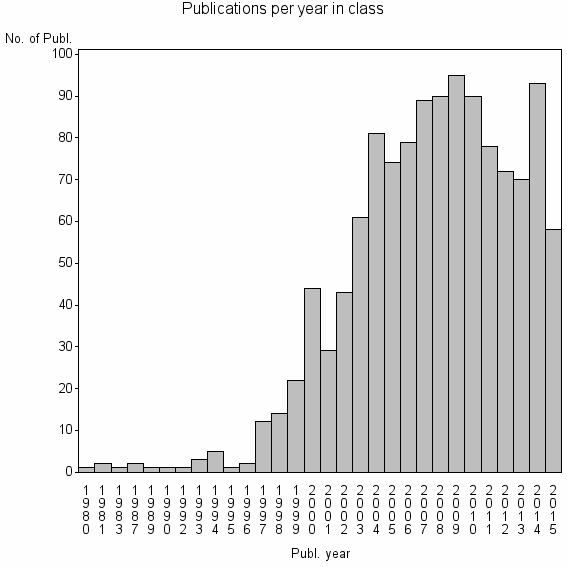
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 8147 | 1214 | 51.5 | 86% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 238 | 19563 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | WHOLE GENOME DUPLICATION | Author keyword | 31 | 41% | 5% | 59 |
| 2 | GENET MOL CELLULAIRE ECT LEVU INTERE | Address | 12 | 75% | 1% | 9 |
| 3 | GENOME DUPLICATION | Author keyword | 12 | 21% | 4% | 52 |
| 4 | GENE DUPLICABILITY | Author keyword | 11 | 100% | 0% | 6 |
| 5 | GENE RETENTION | Author keyword | 11 | 78% | 1% | 7 |
| 6 | CNRSURA1925 | Address | 9 | 83% | 0% | 5 |
| 7 | DOSAGE BALANCE | Author keyword | 9 | 83% | 0% | 5 |
| 8 | INRAUMR216 | Address | 8 | 75% | 0% | 6 |
| 9 | PLANT GENOME M PING | Address | 7 | 17% | 3% | 36 |
| 10 | NEOFUNCTIONALIZATION | Author keyword | 7 | 27% | 2% | 21 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | WHOLE GENOME DUPLICATION | 31 | 41% | 5% | 59 | Search WHOLE+GENOME+DUPLICATION | Search WHOLE+GENOME+DUPLICATION |
| 2 | GENOME DUPLICATION | 12 | 21% | 4% | 52 | Search GENOME+DUPLICATION | Search GENOME+DUPLICATION |
| 3 | GENE DUPLICABILITY | 11 | 100% | 0% | 6 | Search GENE+DUPLICABILITY | Search GENE+DUPLICABILITY |
| 4 | GENE RETENTION | 11 | 78% | 1% | 7 | Search GENE+RETENTION | Search GENE+RETENTION |
| 5 | DOSAGE BALANCE | 9 | 83% | 0% | 5 | Search DOSAGE+BALANCE | Search DOSAGE+BALANCE |
| 6 | NEOFUNCTIONALIZATION | 7 | 27% | 2% | 21 | Search NEOFUNCTIONALIZATION | Search NEOFUNCTIONALIZATION |
| 7 | SUBFUNCTIONALIZATION | 5 | 21% | 2% | 22 | Search SUBFUNCTIONALIZATION | Search SUBFUNCTIONALIZATION |
| 8 | DUPLICATE GENE | 4 | 67% | 0% | 4 | Search DUPLICATE+GENE | Search DUPLICATE+GENE |
| 9 | INTERLOCUS GENE CONVERSION | 4 | 75% | 0% | 3 | Search INTERLOCUS+GENE+CONVERSION | Search INTERLOCUS+GENE+CONVERSION |
| 10 | HOX CLUSTERS | 4 | 56% | 0% | 5 | Search HOX+CLUSTERS | Search HOX+CLUSTERS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SUBFUNCTIONALIZATION | 54 | 57% | 5% | 63 |
| 2 | HOX CLUSTERS | 28 | 47% | 4% | 44 |
| 3 | NEOFUNCTIONALIZATION | 24 | 55% | 2% | 30 |
| 4 | DUPLICABILITY | 23 | 70% | 2% | 19 |
| 5 | DOSAGE SENSITIVITY | 22 | 53% | 2% | 30 |
| 6 | GENOME DUPLICATIONS | 22 | 38% | 4% | 46 |
| 7 | WHOLE GENOME DUPLICATION | 21 | 34% | 4% | 50 |
| 8 | DUPLICATE GENES | 20 | 18% | 8% | 101 |
| 9 | FUNCTIONAL DIVERGENCE | 19 | 24% | 6% | 70 |
| 10 | GENOME DUPLICATION | 17 | 17% | 7% | 88 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The evolution of gene duplications: classifying and distinguishing between models | 2010 | 315 | 74 | 61% |
| Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-Genome, Segmental, or by Transposition | 2009 | 183 | 84 | 54% |
| Turning a hobby into a job: How duplicated genes find new functions | 2008 | 312 | 104 | 38% |
| OPINION The evolutionary significance of ancient genome duplications | 2009 | 232 | 99 | 38% |
| Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time | 2014 | 6 | 74 | 68% |
| Yesterday's polyploids and the mystery of diploidization | 2001 | 371 | 49 | 65% |
| Rapidly evolving fish genomes and teleost diversity | 2008 | 99 | 44 | 59% |
| Expression divergence between duplicate genes | 2005 | 163 | 29 | 79% |
| Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes | 2009 | 98 | 118 | 49% |
| Gene duplication as a driver of plant morphogenetic evolution | 2014 | 8 | 36 | 39% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENET MOL CELLULAIRE ECT LEVU INTERE | 12 | 75% | 0.7% | 9 |
| 2 | CNRSURA1925 | 9 | 83% | 0.4% | 5 |
| 3 | INRAUMR216 | 8 | 75% | 0.5% | 6 |
| 4 | PLANT GENOME M PING | 7 | 17% | 3.0% | 36 |
| 5 | BOT GENET MICROBIOL | 6 | 100% | 0.3% | 4 |
| 6 | BIOINFORMAT BIOL STAT | 5 | 32% | 1.0% | 12 |
| 7 | INRAUMR 216 | 4 | 75% | 0.2% | 3 |
| 8 | UNITE GENET MOL LEVU | 4 | 20% | 1.6% | 19 |
| 9 | CNRSURA 1925 | 3 | 57% | 0.3% | 4 |
| 10 | BIOL CELLULAIRE LEVURE | 3 | 38% | 0.5% | 6 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000213292 | ASCOBOLUS//EVOLUTION OF PLOIDY//MULTIGENE ARRAYS |
| 2 | 0.0000204802 | SEX BIASED GENE EXPRESSION//SEXUAL ANTAGONISM//INTRALOCUS SEXUAL CONFLICT |
| 3 | 0.0000183623 | PROCESSED PSEUDOGENES//PHYSICAL AND PSYCHOLOGICAL STRESSES//DUPLICATED PSEUDOGENES |
| 4 | 0.0000169140 | IROQUOIS//CONSERVED NONCODING ELEMENTS//ENERGY ENVIRONM BIOL COMP |
| 5 | 0.0000156400 | RICE GENOME PROGRAM//CROP DESIGN//GENOME IL |
| 6 | 0.0000153292 | CIRCOS//BIODADOS//ORTHOLOGY |
| 7 | 0.0000143505 | GENOME REARRANGEMENTS//SORTING BY REVERSALS//BREAKPOINT GRAPH |
| 8 | 0.0000133856 | POLYPLOIDY//AUTOPOLYPLOIDY//ALLOPOLYPLOIDY |
| 9 | 0.0000120907 | BIDIRECTIONAL PROMOTER//NEIGHBORING GENES//GENOME NETWORK PROJECT CORE GRPGENOM SCI |
| 10 | 0.0000075425 | AVIDA//EVOLVABILITY//DIGITAL EVOLUTION |