Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
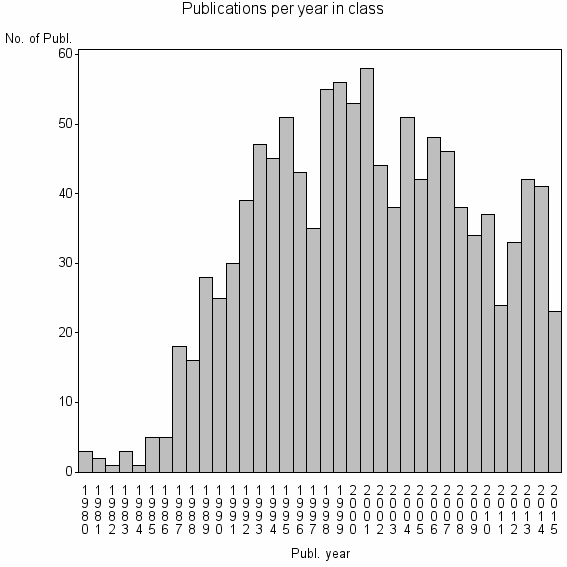
| 8680 | 1160 | 43.3 | 79% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | STAPHYLOCOCCAL NUCLEASE | Author keyword | 22 | 38% | 4% | 47 |
| 2 | THERMODYNAMIC MOLECULAR SWITCH | Author keyword | 8 | 100% | 0% | 5 |
| 3 | PROTEIN THERMOSTABILITY | Author keyword | 7 | 28% | 2% | 21 |
| 4 | INNATE TEMPERATURE INVARIANT ENTHALPY | Author keyword | 6 | 100% | 0% | 4 |
| 5 | THERMOPHILIC PROTEIN | Author keyword | 5 | 41% | 1% | 9 |
| 6 | THERMOPHILIC PROTEINS | Author keyword | 4 | 38% | 1% | 8 |
| 7 | LOCAL STRUCTURAL ENTROPY | Author keyword | 3 | 100% | 0% | 3 |
| 8 | MESOPHILIC PROTEINS | Author keyword | 3 | 100% | 0% | 3 |
| 9 | PLANCK BENZINGER THERMAL WORK FUNCTION | Author keyword | 3 | 100% | 0% | 3 |
| 10 | UNFOLDING FREE ENERGY CHANGE | Author keyword | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | HYPERTHERMOPHILIC PROTEINS | 35 | 58% | 3% | 40 |
| 2 | THERMOPHILIC PROTEINS | 32 | 48% | 4% | 49 |
| 3 | THERMAL WORK FUNCTION | 23 | 100% | 1% | 10 |
| 4 | BACTERIOPHAGE T4 LYSOZYME | 21 | 29% | 5% | 62 |
| 5 | THERMOSTABILITY | 21 | 12% | 14% | 158 |
| 6 | SALT BRIDGES | 18 | 19% | 8% | 88 |
| 7 | T4 LYSOZYME | 16 | 14% | 10% | 111 |
| 8 | MESOPHILIC PROTEINS | 15 | 68% | 1% | 13 |
| 9 | CAVITY CREATING MUTATIONS | 12 | 30% | 3% | 34 |
| 10 | COLD SHOCK PROTEIN | 12 | 19% | 5% | 56 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Lessons from the lysozyme of phage T4 | 2010 | 56 | 62 | 48% |
| Lessons in stability from thermophilic proteins | 2006 | 132 | 74 | 46% |
| Forces stabilizing proteins | 2014 | 8 | 82 | 38% |
| Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins | 2008 | 43 | 73 | 49% |
| Thermophilic proteins: insight and perspective from in silico experiments | 2012 | 16 | 57 | 54% |
| STUDIES ON PROTEIN STABILITY WITH T4 LYSOZYME | 1995 | 257 | 27 | 81% |
| Contribution of surface salt bridges to protein stability: Guidelines for protein engineering | 2003 | 125 | 92 | 45% |
| STRUCTURAL AND GENETIC-ANALYSIS OF PROTEIN STABILITY | 1993 | 385 | 41 | 59% |
| Forces contributing to the conformational stability of proteins | 1996 | 369 | 34 | 44% |
| Structural features of thermozymes | 2005 | 86 | 36 | 39% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ADJUNCT BIOL | 1 | 50% | 0.2% | 2 |
| 2 | FREDERICK CANC DEV EXPT COMPUTAT | 1 | 50% | 0.2% | 2 |
| 3 | UNITE BIOINFORMAT GENOM STRUCT | 1 | 23% | 0.3% | 3 |
| 4 | ADV ENVIRONM GENOMICS | 1 | 50% | 0.1% | 1 |
| 5 | AG PARASKEVI ATTIKIS | 1 | 50% | 0.1% | 1 |
| 6 | BAYREUTH ZENTRUM MOL BIOWISSEN | 1 | 50% | 0.1% | 1 |
| 7 | BIOL CHEM CHEM BIOL | 1 | 50% | 0.1% | 1 |
| 8 | EA3763 | 1 | 50% | 0.1% | 1 |
| 9 | HEMATOL MED ONCOL PATHOL | 1 | 50% | 0.1% | 1 |
| 10 | HUA QIAO UNIV | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000194712 | PSYCHROPHILE//COLD ADAPTATION//PSYCHROPHILES |
| 2 | 0.0000125181 | THERMOLYSIN//HALOPHILICITY//NEUTRAL PROTEASE |
| 3 | 0.0000117361 | COMPUTATIONAL PROTEIN DESIGN//DEAD END ELIMINATION//COMPUTATIONAL ENZYME DESIGN |
| 4 | 0.0000116356 | PROTEIN HYDRATION//INTERNAL SOLVENT//SOLVENT MODELING |
| 5 | 0.0000112861 | POISSON BOLTZMANN EQUATION//POISSON BOLTZMANN//BIOMOLECULAR ELECTROSTATICS |
| 6 | 0.0000108500 | BPTI//BAKER CHEM CHEM BIOL//SCRAMBLED PROTEINS |
| 7 | 0.0000103783 | BETA HAIRPIN//SIDE CHAIN INTERACTIONS//TRP CAGE |
| 8 | 0.0000102197 | TRYPTOPHAN SYNTHASE//INDOLE 3 GLYCEROL PHOSPHATE SYNTHASE//SECT ENZYME STRUCT FUNCT |
| 9 | 0.0000095444 | PROTEIN FOLDING//PHI VALUE//CONTACT ORDER |
| 10 | 0.0000095288 | PROTEIN MELTING DOMAINS//THERMAL SHIFT ASSAYS//THERMOFLUOR |