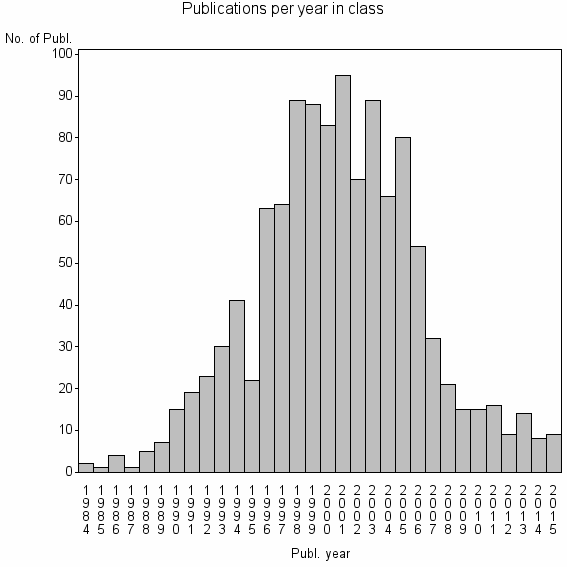
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 8768 | 1150 | 24.5 | 74% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 238 | 19563 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EMBL OUTSTN | Address | 11 | 21% | 4% | 48 |
| 2 | PROT INFORMAT OURCE | Address | 5 | 44% | 1% | 8 |
| 3 | HLTH SCI TECHNOL ICIR | Address | 4 | 75% | 0% | 3 |
| 4 | J AN INT PROT INFORMAT DATABASE | Address | 3 | 100% | 0% | 3 |
| 5 | PROTEIN SEQUENCE DATABASE | Author keyword | 3 | 45% | 0% | 5 |
| 6 | EUROPEAN BIOINFORMAT | Address | 3 | 10% | 2% | 26 |
| 7 | OXFORD MOL GRP | Address | 2 | 67% | 0% | 2 |
| 8 | BOLOGNA BIOCOMP GRP | Address | 2 | 50% | 0% | 3 |
| 9 | EPIDEMIOL BIOMATH | Address | 2 | 43% | 0% | 3 |
| 10 | INTERPRO | Author keyword | 1 | 38% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN SEQUENCE DATABASE | 3 | 45% | 0% | 5 | Search PROTEIN+SEQUENCE+DATABASE | Search PROTEIN+SEQUENCE+DATABASE |
| 2 | INTERPRO | 1 | 38% | 0% | 3 | Search INTERPRO | Search INTERPRO |
| 3 | BIOMOLECULAR DATABASE | 1 | 50% | 0% | 2 | Search BIOMOLECULAR+DATABASE | Search BIOMOLECULAR+DATABASE |
| 4 | BLAST SOFTWARE | 1 | 100% | 0% | 2 | Search BLAST+SOFTWARE | Search BLAST+SOFTWARE |
| 5 | CLUSTR | 1 | 100% | 0% | 2 | Search CLUSTR | Search CLUSTR |
| 6 | MOLECULAR BIOLOGY DATABASE | 1 | 100% | 0% | 2 | Search MOLECULAR+BIOLOGY+DATABASE | Search MOLECULAR+BIOLOGY+DATABASE |
| 7 | SUPERFAMILY CLASSIFICATION | 1 | 50% | 0% | 2 | Search SUPERFAMILY+CLASSIFICATION | Search SUPERFAMILY+CLASSIFICATION |
| 8 | MOLECULAR BIOLOGY DATABASES | 1 | 33% | 0% | 3 | Search MOLECULAR+BIOLOGY+DATABASES | Search MOLECULAR+BIOLOGY+DATABASES |
| 9 | CROSS GENOME COMPARISON | 1 | 40% | 0% | 2 | Search CROSS+GENOME+COMPARISON | Search CROSS+GENOME+COMPARISON |
| 10 | PROTEIN SEQUENCE CLUSTERING | 1 | 40% | 0% | 2 | Search PROTEIN+SEQUENCE+CLUSTERING | Search PROTEIN+SEQUENCE+CLUSTERING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | TREMBL | 13 | 51% | 2% | 18 |
| 2 | PRODOM | 9 | 83% | 0% | 5 |
| 3 | CLUSTR | 7 | 67% | 1% | 6 |
| 4 | SEQUENCE DATABASE | 6 | 18% | 3% | 30 |
| 5 | PROSITE | 6 | 41% | 1% | 11 |
| 6 | SWISS PROT | 5 | 22% | 2% | 20 |
| 7 | DATABASE CLASSIFICATION | 4 | 67% | 0% | 4 |
| 8 | EBI SRS SERVER | 4 | 75% | 0% | 3 |
| 9 | STRAIN RD KW20 | 4 | 75% | 0% | 3 |
| 10 | PEDANT | 3 | 60% | 0% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| In silico characterization of proteins: UniProt, InterPro and Integr8 | 2008 | 35 | 42 | 31% |
| Protein sequence databases | 2004 | 92 | 29 | 52% |
| Protein data bank archives of three-dimensional macromolecular structures | 1997 | 166 | 9 | 56% |
| Genome Reviews: Standardizing content and representation of information about complete genomes | 2006 | 16 | 9 | 44% |
| Archiving for Psychologists: Suggestions for Organizing, Documenting, Preserving, and Protecting Computer Files | 2011 | 0 | 2 | 100% |
| The human proteomics initiative (HPI) | 2001 | 45 | 14 | 71% |
| XML, bioinformatics and data integration | 2001 | 112 | 21 | 38% |
| Automatic annotation of protein function | 2005 | 59 | 72 | 29% |
| ISYS (Integrated SYStem): a platform for integrating heterogeneous bioinformatic resources | 2002 | 1 | 2 | 100% |
| Molecular biology databases: today and tomorrow | 2001 | 3 | 11 | 100% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EMBL OUTSTN | 11 | 21% | 4.2% | 48 |
| 2 | PROT INFORMAT OURCE | 5 | 44% | 0.7% | 8 |
| 3 | HLTH SCI TECHNOL ICIR | 4 | 75% | 0.3% | 3 |
| 4 | J AN INT PROT INFORMAT DATABASE | 3 | 100% | 0.3% | 3 |
| 5 | EUROPEAN BIOINFORMAT | 3 | 10% | 2.3% | 26 |
| 6 | OXFORD MOL GRP | 2 | 67% | 0.2% | 2 |
| 7 | BOLOGNA BIOCOMP GRP | 2 | 50% | 0.3% | 3 |
| 8 | EPIDEMIOL BIOMATH | 2 | 43% | 0.3% | 3 |
| 9 | MEMOREC | 1 | 100% | 0.2% | 2 |
| 10 | SUDARSKY COMPUTAT BIOL | 1 | 20% | 0.5% | 6 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000193154 | PROTEIN STRUCTURE PREDICTION//FOLD RECOGNITION//CASP |
| 2 | 0.0000158029 | GENE PREDICTION//GENE FINDING//GENE RECOGNITION |
| 3 | 0.0000150811 | CIRCOS//BIODADOS//ORTHOLOGY |
| 4 | 0.0000120002 | COMPLEX SCI SOFTWARE//3D ZERNIKE DESCRIPTOR//PROTEIN SURFACE SHAPE |
| 5 | 0.0000119699 | JOURNAL OF BIOMEDICAL SEMANTICS//ZFIN//ZEBRAFISH INFORMAT NETWORK |
| 6 | 0.0000115749 | EVOLUTIONARY BIOINFORMAT//CLOSED LOOPS//SKEW PAIRS |
| 7 | 0.0000097411 | MKIAA//SMART CDNA LIBRARY//CDNA SEQUENCING |
| 8 | 0.0000081109 | SECONDARY STRUCTURE PREDICTION//PROTEIN SECONDARY STRUCTURE PREDICTION//COMPOUND PYRAMID MODEL |
| 9 | 0.0000081099 | AMBIGRAM//DESK TOP MOLECULAR MODELING//PARTIAL DIGEST |
| 10 | 0.0000080636 | BIOMAG BANK//PDBJ//RCSB PROT DATA BANK |