Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
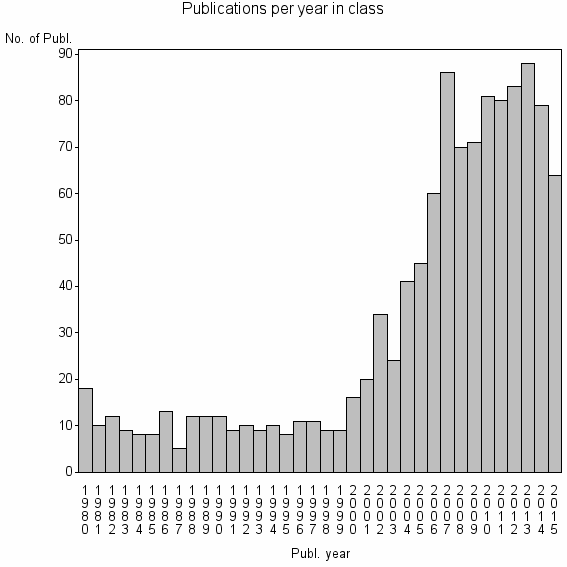
| 8808 | 1147 | 47.1 | 73% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 623 | 13179 | BASE EXCISION REPAIR//DNA GLYCOSYLASE//URACIL DNA GLYCOSYLASE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | DNA PHOTOPHYSICS | Author keyword | 15 | 88% | 1% | 7 |
| 2 | BIO NANOSYST SCI | Address | 14 | 100% | 1% | 7 |
| 3 | DNA PHOTOCHEMISTRY | Author keyword | 9 | 67% | 1% | 8 |
| 4 | SEMICLASSICAL DYNAMICS SIMULATION | Author keyword | 8 | 75% | 1% | 6 |
| 5 | FRANCIS PERRINURA 2453 | Address | 7 | 36% | 1% | 15 |
| 6 | NUCLEOBASE ANALOGUES | Author keyword | 6 | 80% | 0% | 4 |
| 7 | BONDED EXCIMER | Author keyword | 6 | 100% | 0% | 4 |
| 8 | ULTRAFAST RADIATIONLESS DECAY | Author keyword | 6 | 100% | 0% | 4 |
| 9 | N PHOTOTECH | Address | 4 | 67% | 0% | 4 |
| 10 | FRANCIS PERRIN | Address | 4 | 14% | 2% | 28 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ULTRAFAST INTERNAL CONVERSION | 146 | 59% | 14% | 163 |
| 2 | ULTRAFAST DECAY | 75 | 91% | 3% | 31 |
| 3 | PYRIMIDINE NUCLEOBASES | 69 | 96% | 2% | 22 |
| 4 | JET COOLED ADENINE | 61 | 78% | 3% | 40 |
| 5 | ELECTRONIC RELAXATION DYNAMICS | 50 | 71% | 4% | 41 |
| 6 | EXCITED STATE DYNAMICS | 49 | 19% | 20% | 229 |
| 7 | A T DNA | 49 | 94% | 1% | 17 |
| 8 | RNA BASES | 40 | 49% | 5% | 59 |
| 9 | NUCLEIC ACID BASES | 38 | 16% | 19% | 215 |
| 10 | FLUORESCENCE UP CONVERSION | 38 | 40% | 7% | 75 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Excitation of Nucleobases from a Computational Perspective I: Reaction Paths | 2015 | 11 | 132 | 70% |
| Excited state dynamics of DNA bases | 2013 | 54 | 229 | 79% |
| DNA Excited-State Dynamics: From Single Bases to the Double Helix | 2009 | 270 | 97 | 80% |
| Electronic Excitation Processes in Single-Strand and Double-Strand DNA: A Computational Approach | 2015 | 4 | 230 | 77% |
| Excited States Behavior of Nucleobases in Solution: Insights from Computational Studies | 2015 | 4 | 122 | 62% |
| Excitation of Nucleobases from a Computational Perspective II: Dynamics | 2015 | 4 | 91 | 62% |
| Are the five natural DNA/RNA base monomers a good choice from natural selection? A photochemical perspective | 2009 | 108 | 87 | 76% |
| Ultrafast excited-state dynamics in nucleic acids | 2004 | 640 | 247 | 44% |
| Progress and Challenges in the Calculation of Electronic Excited States | 2012 | 100 | 302 | 24% |
| A doorway state leads to photostability or triplet photodamage in thymine DNA | 2008 | 82 | 81 | 70% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIO NANOSYST SCI | 14 | 100% | 0.6% | 7 |
| 2 | FRANCIS PERRINURA 2453 | 7 | 36% | 1.3% | 15 |
| 3 | N PHOTOTECH | 4 | 67% | 0.3% | 4 |
| 4 | FRANCIS PERRIN | 4 | 14% | 2.4% | 28 |
| 5 | URA 2453 | 3 | 21% | 1.2% | 14 |
| 6 | COMPUTAT MOL MODELING STRUCT INTERACT | 2 | 50% | 0.3% | 3 |
| 7 | PHYS CHEM ELEKTROCHEM 1 | 2 | 17% | 1.0% | 11 |
| 8 | THEORET CHEM PROGRAM | 1 | 38% | 0.3% | 3 |
| 9 | CEA DSM DRECAM SPAM FRANCIS PERRIN | 1 | 100% | 0.2% | 2 |
| 10 | N PHOTTECH | 1 | 100% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000217459 | MOL QUANTUM BIOPHYS//MOL BIOL BIOTECHNOL BIOPHYS//THE DOUBLE PROTON TRANSFER |
| 2 | 0.0000150890 | VIBRATION ROTATIONAL CONSTANTS//CI CALCULATIONS//SPECT ELECT DIFFUSES |
| 3 | 0.0000128214 | NANOSECOND UV PHOTOLYSIS//2 METHOXYCYTOSINE//ACETONE SENSITIZATION |
| 4 | 0.0000099801 | S ESPECT BIOESPECT//ESPE OSCOPIA BIOESPE OSCOPIA//S ESPE OSCOPIA BIOESPE OSCOPIA |
| 5 | 0.0000099658 | DNA ELECTRON TRANSFER//DNA CHARGE TRANSPORT//SCI IND SANKEN |
| 6 | 0.0000085220 | TIME DEPENDENT DENSITY FUNCTIONAL THEORY//GRP CHIM PHYS THEOR STRUCT//CHIM THEOR PL |
| 7 | 0.0000084522 | PHOTOLYASE//DNA PHOTOLYASE//PHOTOLYASES |
| 8 | 0.0000080859 | GRP CANADIAN HLTH RADIAT SCI//GRP SCI RADIAT//HBEREICH CHEM BIOL 2 |
| 9 | 0.0000075281 | 4 THIOTHYMIDINE//THIOTHYMIDINE//NITROSAMINE INDUCED CANC GRP |
| 10 | 0.0000074062 | GENOME RADIAT BIOL//DNA PHOTOLESIONS//DNA PHOTOPRODUCTS |