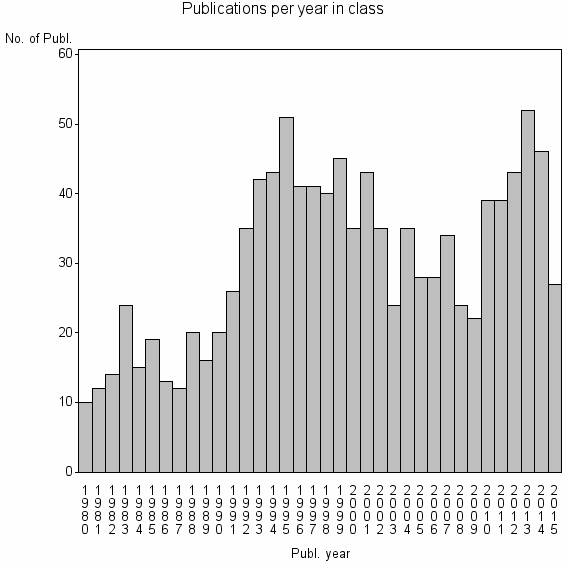
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 9366 | 1093 | 40.3 | 77% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 483 | 14947 | GANGLIOSIDES//SIALYLTRANSFERASE//SIALIC ACID |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | TN ANTIGEN | Author keyword | 44 | 54% | 5% | 57 |
| 2 | SIALYL TN | Author keyword | 27 | 55% | 3% | 34 |
| 3 | SIALYL TN ANTIGEN | Author keyword | 26 | 67% | 2% | 24 |
| 4 | THOMSEN FRIEDENREICH ANTIGEN | Author keyword | 24 | 50% | 3% | 34 |
| 5 | O GLYCOSYLATION | Author keyword | 22 | 21% | 9% | 94 |
| 6 | PPGALNAC T | Author keyword | 20 | 100% | 1% | 9 |
| 7 | HELIX POMATIA AGGLUTININ | Author keyword | 19 | 74% | 1% | 14 |
| 8 | UDP GALNAC POLYPEPTIDE N ACETYLGALACTOSAMINYLTRANSFERASE | Author keyword | 17 | 100% | 1% | 8 |
| 9 | DEV GLYCOBIOL UNIT | Address | 15 | 73% | 1% | 11 |
| 10 | GALNAC TRANSFERASE | Author keyword | 14 | 60% | 1% | 15 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TN ANTIGEN | 44 | 54% | 5% | 57 | Search TN+ANTIGEN | Search TN+ANTIGEN |
| 2 | SIALYL TN | 27 | 55% | 3% | 34 | Search SIALYL+TN | Search SIALYL+TN |
| 3 | SIALYL TN ANTIGEN | 26 | 67% | 2% | 24 | Search SIALYL+TN+ANTIGEN | Search SIALYL+TN+ANTIGEN |
| 4 | THOMSEN FRIEDENREICH ANTIGEN | 24 | 50% | 3% | 34 | Search THOMSEN+FRIEDENREICH+ANTIGEN | Search THOMSEN+FRIEDENREICH+ANTIGEN |
| 5 | O GLYCOSYLATION | 22 | 21% | 9% | 94 | Search O+GLYCOSYLATION | Search O+GLYCOSYLATION |
| 6 | PPGALNAC T | 20 | 100% | 1% | 9 | Search PPGALNAC+T | Search PPGALNAC+T |
| 7 | HELIX POMATIA AGGLUTININ | 19 | 74% | 1% | 14 | Search HELIX+POMATIA+AGGLUTININ | Search HELIX+POMATIA+AGGLUTININ |
| 8 | UDP GALNAC POLYPEPTIDE N ACETYLGALACTOSAMINYLTRANSFERASE | 17 | 100% | 1% | 8 | Search UDP+GALNAC++POLYPEPTIDE+N+ACETYLGALACTOSAMINYLTRANSFERASE | Search UDP+GALNAC++POLYPEPTIDE+N+ACETYLGALACTOSAMINYLTRANSFERASE |
| 9 | GALNAC TRANSFERASE | 14 | 60% | 1% | 15 | Search GALNAC+TRANSFERASE | Search GALNAC+TRANSFERASE |
| 10 | GALNAC T3 | 12 | 86% | 1% | 6 | Search GALNAC+T3 | Search GALNAC+T3 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ALPHA D GALACTOSAMINE | 130 | 71% | 10% | 104 |
| 2 | SIALOSYL TN | 112 | 56% | 13% | 138 |
| 3 | ACETYL D GALACTOSAMINE | 51 | 50% | 7% | 74 |
| 4 | POLYPEPTIDE N ACETYLGALACTOSAMINYLTRANSFERASE | 50 | 42% | 8% | 91 |
| 5 | N ACETYLGALACTOSAMINYLTRANSFERASE FAMILY | 49 | 94% | 2% | 17 |
| 6 | UDP GALNAC | 49 | 50% | 6% | 71 |
| 7 | BOVINE UDP GALNAC | 31 | 76% | 2% | 22 |
| 8 | HELIX POMATIA AGGLUTININ | 27 | 65% | 2% | 26 |
| 9 | SIALYL TN ANTIGEN | 20 | 50% | 3% | 29 |
| 10 | GALNAC | 17 | 44% | 3% | 29 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family | 2012 | 75 | 161 | 51% |
| Mucin-type O-Glycosylation during Development | 2013 | 28 | 72 | 63% |
| Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy | 1997 | 253 | 40 | 58% |
| All in the family: the UDP-GalNAc : polypeptide N-acetylgalactosaminyltransferases | 2003 | 170 | 144 | 54% |
| Alterations in glycosylation as biomarkers for cancer detection | 2010 | 103 | 130 | 38% |
| Cell surface protein glycosylation in cancer | 2014 | 29 | 188 | 14% |
| Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions | 2006 | 190 | 54 | 43% |
| Location, location, location: new insights into O-GaINAc protein glycosylation | 2011 | 40 | 100 | 50% |
| The Tn Antigen-Structural Simplicity and Biological Complexity | 2011 | 63 | 281 | 31% |
| NetOglyc: Prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility | 1998 | 349 | 114 | 28% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DEV GLYCOBIOL UNIT | 15 | 73% | 1.0% | 11 |
| 2 | COPENHAGEN GLYC | 7 | 29% | 2.0% | 22 |
| 3 | WA BERNBAUM CYST FIBROSIS | 7 | 53% | 0.8% | 9 |
| 4 | GLYCOGENE FUNCT TEAM | 5 | 29% | 1.3% | 14 |
| 5 | ONCOL BASICA | 4 | 67% | 0.4% | 4 |
| 6 | POSTGENOME SCI TECHNOL | 4 | 75% | 0.3% | 3 |
| 7 | GLICOBIOL INMUNOL TUMORAL | 3 | 57% | 0.4% | 4 |
| 8 | HEATHER M BLIGH CANC S | 3 | 100% | 0.3% | 3 |
| 9 | MED SURG BREAST CANC GRP | 3 | 100% | 0.3% | 3 |
| 10 | SECT BIOL CHEM | 3 | 16% | 1.5% | 16 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000226564 | TUMOR VACCINOL//GLYCOPEPTIDES//CARL S MARVEL S |
| 2 | 0.0000163055 | MUC1//MUC1 MUCIN//MUC 1 |
| 3 | 0.0000143310 | SIALYLTRANSFERASE//ST6GAL I//BETA GALACTOSIDE ALPHA 2 6 SIALYLTRANSFERASE |
| 4 | 0.0000139238 | LECTIN HISTOCHEMISTRY//TRANSITIONAL MUCOSA//ORGANOID DIFFERENTIATION |
| 5 | 0.0000131996 | T ACTIVATION//POLYAGGLUTINATION//ERYTHROCYTE CHARGE |
| 6 | 0.0000122692 | MUC4//MUCIN//MUC2 |
| 7 | 0.0000117719 | FUCOSYLTRANSFERASE//LEWIS Y ANTIGEN//FUT2 |
| 8 | 0.0000112807 | N ACETYLGLUCOSAMINYLTRANSFERASE V//GNT V//GNT III |
| 9 | 0.0000103701 | GLYCOIMMUNOCHEM//GLYCOPROTEIN BINDING//CARBOHYDRATE SPECIFICITIES |
| 10 | 0.0000093815 | TOXOPLASMA LYSATE ANTIGEN//OBIOPEPTIDE//L5178Y R LYMPHOMA |