Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
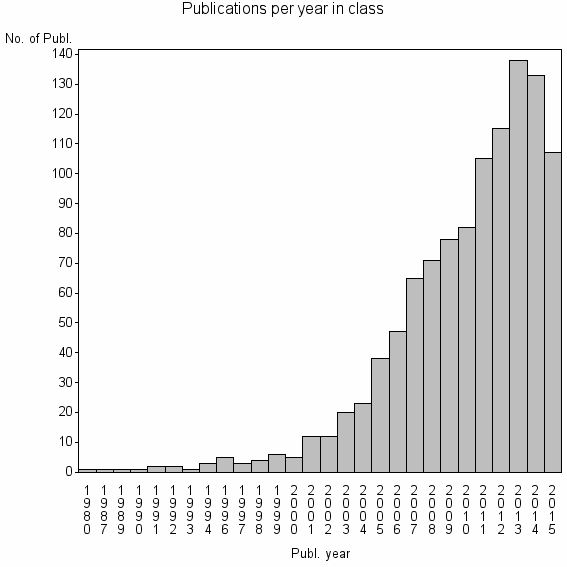
| 9520 | 1081 | 45.0 | 81% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1176 | 8954 | METABONOMICS//METABOLOMICS//METABOLIC PROFILING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | METABOLITE ANNOTATION | Author keyword | 14 | 100% | 1% | 7 |
| 2 | SCRIPPS METABOL MASS SPE OMETRY | Address | 11 | 69% | 1% | 9 |
| 3 | METABOLOMICS | Journal | 9 | 10% | 7% | 78 |
| 4 | METABOLITE PROFILING | Author keyword | 7 | 11% | 5% | 57 |
| 5 | RETENTION TIME ALIGNMENT | Author keyword | 7 | 67% | 1% | 6 |
| 6 | PROF L WILLMITZER | Address | 4 | 67% | 0% | 4 |
| 7 | PLANT METABOLOMICS | Author keyword | 4 | 38% | 1% | 9 |
| 8 | CORRELATION OPTIMISED WARPING | Author keyword | 3 | 100% | 0% | 3 |
| 9 | KNAPSACK FAMILY | Author keyword | 3 | 100% | 0% | 3 |
| 10 | PARAMETRIC TIME WARPING | Author keyword | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | METABOLITE ANNOTATION | 14 | 100% | 1% | 7 | Search METABOLITE+ANNOTATION | Search METABOLITE+ANNOTATION |
| 2 | METABOLITE PROFILING | 7 | 11% | 5% | 57 | Search METABOLITE+PROFILING | Search METABOLITE+PROFILING |
| 3 | RETENTION TIME ALIGNMENT | 7 | 67% | 1% | 6 | Search RETENTION+TIME+ALIGNMENT | Search RETENTION+TIME+ALIGNMENT |
| 4 | PLANT METABOLOMICS | 4 | 38% | 1% | 9 | Search PLANT+METABOLOMICS | Search PLANT+METABOLOMICS |
| 5 | CORRELATION OPTIMISED WARPING | 3 | 100% | 0% | 3 | Search CORRELATION+OPTIMISED+WARPING | Search CORRELATION+OPTIMISED+WARPING |
| 6 | KNAPSACK FAMILY | 3 | 100% | 0% | 3 | Search KNAPSACK+FAMILY | Search KNAPSACK+FAMILY |
| 7 | PARAMETRIC TIME WARPING | 3 | 100% | 0% | 3 | Search PARAMETRIC+TIME+WARPING | Search PARAMETRIC+TIME+WARPING |
| 8 | SPECIES METABOLITE RELATIONSHIP | 3 | 100% | 0% | 3 | Search SPECIES+METABOLITE+RELATIONSHIP | Search SPECIES+METABOLITE+RELATIONSHIP |
| 9 | SPECTRUM CLEAN UP | 3 | 100% | 0% | 3 | Search SPECTRUM+CLEAN+UP | Search SPECTRUM+CLEAN+UP |
| 10 | MQTL | 2 | 44% | 0% | 4 | Search MQTL | Search MQTL |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PLANT METABOLOMICS | 32 | 38% | 6% | 68 |
| 2 | HIGH RESOLUTION LC MS | 18 | 83% | 1% | 10 |
| 3 | COMPOUND IDENTIFICATION | 17 | 45% | 3% | 28 |
| 4 | LC MS DATA | 16 | 56% | 2% | 19 |
| 5 | COMPONENT DETECTION | 15 | 88% | 1% | 7 |
| 6 | MASS SPECTROMETRY DATA | 12 | 16% | 6% | 68 |
| 7 | FRAGMENTATION TREES | 11 | 78% | 1% | 7 |
| 8 | PEAK ALIGNMENT | 10 | 29% | 3% | 29 |
| 9 | MOLECULAR PROFILE DATA | 7 | 46% | 1% | 12 |
| 10 | METABOLITE IDENTIFICATION | 7 | 21% | 3% | 31 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | METABOLOMICS | 9 | 10% | 7% | 78 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| LC-MS alignment in theory and practice: a comprehensive algorithmic review | 2015 | 4 | 59 | 58% |
| Computational mass spectrometry for small-molecule fragmentation | 2014 | 10 | 44 | 75% |
| High-throughput discovery metabolomics | 2015 | 5 | 39 | 26% |
| New kids on the block: novel informatics methods for natural product discovery | 2014 | 8 | 34 | 62% |
| Current approaches and challenges for the metabolite profiling of complex natural extracts | 2015 | 7 | 283 | 17% |
| Integrated metabolomics for abiotic stress responses in plants | 2015 | 4 | 66 | 27% |
| Metabolomic-Based Strategies for Anti-Parasite Drug Discovery | 2015 | 5 | 91 | 21% |
| Data processing for mass spectrometry-based metabolomics | 2007 | 208 | 75 | 36% |
| Computational mass spectrometry for small molecules | 2013 | 17 | 207 | 45% |
| Current metabolomics: Technological advances | 2013 | 23 | 66 | 35% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SCRIPPS METABOL MASS SPE OMETRY | 11 | 69% | 0.8% | 9 |
| 2 | PROF L WILLMITZER | 4 | 67% | 0.4% | 4 |
| 3 | ST S DEV BIOL | 3 | 16% | 1.7% | 18 |
| 4 | BVI IFR103 | 2 | 67% | 0.2% | 2 |
| 5 | CHIM BIOL ORGAN STRUCT | 2 | 67% | 0.2% | 2 |
| 6 | GRONINGEN BIOINFORMAT | 2 | 10% | 1.6% | 17 |
| 7 | BIOSYSTEMETR GRP | 2 | 43% | 0.3% | 3 |
| 8 | GOFORSYS | 2 | 33% | 0.4% | 4 |
| 9 | B2S | 1 | 100% | 0.2% | 2 |
| 10 | IPCM CNRS UMR 7201 | 1 | 100% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000229900 | METABONOMICS//METABOLOMICS//METABOLIC PROFILING |
| 2 | 0.0000198729 | SECT METABOL//CARBOHYDRATE NMR//NMR BASED METABOLOMICS |
| 3 | 0.0000192914 | METABOLIC FLUX ANALYSIS//RAPID SAMPLING//INTRACELLULAR METABOLITES |
| 4 | 0.0000177210 | ANOVA PCA//FUZZY RULE BUILDING EXPERT SYSTEM//INTELLIGENT CHEM RUMENTAT |
| 5 | 0.0000169974 | DISCOVERY ANALYT CHEM CHEM TECHNOL//LC MS TECHNOL DEV//SHANGHAI MASS SPE OMETRY |
| 6 | 0.0000117622 | CIS MOTIF//ARABIDOPSIS T87 CELL LINE//GENE ATLAS |
| 7 | 0.0000081468 | DEMENTIA SERV//FIRST EPISODE NEUROLEPTIC NAIVE//SECONDARY FRACTIONATION |
| 8 | 0.0000080762 | CHROMATOGRAPHIC FINGERPRINT//RADIX ISATIDIS//TKM CONVERGING |
| 9 | 0.0000080591 | ECOTOXICOGENOMICS//ENVIRONMENTAL METABOLOMICS//ENVIRONM PROTE |
| 10 | 0.0000075632 | COMPREHENSIVE TWO DIMENSIONAL GAS CHROMATOGRAPHY//GCXGC//COMPREHENSIVE TWO DIMENSIONAL |