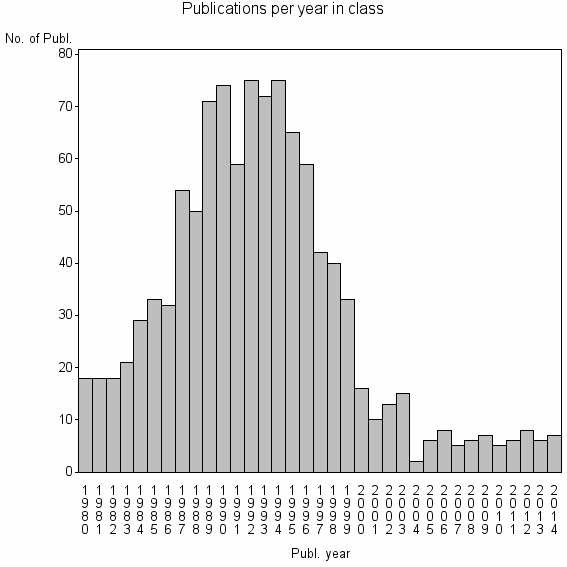
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 9752 | 1058 | 34.2 | 64% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 583 | 13522 | JOURNAL OF MAGNETIC RESONANCE//JOURNAL OF BIOMOLECULAR NMR//RESIDUAL DIPOLAR COUPLINGS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | 3J COUPLING CONSTANTS | Author keyword | 6 | 71% | 0% | 5 |
| 2 | DISTANCE GEOMETRY | Author keyword | 5 | 14% | 3% | 35 |
| 3 | TIME AVERAGED RESTRAINTS | Author keyword | 4 | 75% | 0% | 3 |
| 4 | COMPLETE RELAXATION MATRIX ANALYSIS | Author keyword | 4 | 56% | 0% | 5 |
| 5 | OFF RESONANCE ROESY | Author keyword | 3 | 57% | 0% | 4 |
| 6 | 3J COUPLING CONSTANT | Author keyword | 3 | 100% | 0% | 3 |
| 7 | NOE UPPER BOUNDS | Author keyword | 3 | 100% | 0% | 3 |
| 8 | SAMPLING CONFORMATION SPACE | Author keyword | 3 | 100% | 0% | 3 |
| 9 | NOE RESTRAINTS | Author keyword | 3 | 60% | 0% | 3 |
| 10 | RELAXATION MATRIX | Author keyword | 2 | 36% | 0% | 5 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | OVERHAUSER EFFECT SPECTRA | 75 | 60% | 8% | 81 |
| 2 | INTERPROTON DISTANCES | 55 | 87% | 3% | 27 |
| 3 | RELAXATION MATRIX ANALYSIS | 19 | 36% | 4% | 42 |
| 4 | RATE MATRIX ANALYSIS | 14 | 100% | 1% | 7 |
| 5 | NOE INTENSITIES | 11 | 78% | 1% | 7 |
| 6 | RELAXATION MATRIX | 11 | 78% | 1% | 7 |
| 7 | RELAXATION MATRIX APPROACH | 10 | 31% | 3% | 27 |
| 8 | SYNCHRONOUS NUTATION | 9 | 64% | 1% | 9 |
| 9 | CROSS PEAK INTENSITIES | 9 | 83% | 0% | 5 |
| 10 | DIMENSIONAL NOE SPECTRA | 9 | 83% | 0% | 5 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Ambiguous NOEs and automated NOE assignment | 1998 | 166 | 135 | 32% |
| The nuclear Overhauser effect from a quantitative perspective | 2014 | 3 | 200 | 48% |
| RECENT DEVELOPMENTS IN TRANSFERRED NOE METHODS | 1994 | 246 | 148 | 41% |
| Structure calculation of biological macromolecules from NMR data | 1998 | 90 | 150 | 41% |
| AN EVALUATION OF COMPUTATIONAL STRATEGIES FOR USE IN THE DETERMINATION OF PROTEIN-STRUCTURE FROM DISTANCE CONSTRAINTS OBTAINED BY NUCLEAR-MAGNETIC-RESONANCE | 1991 | 349 | 63 | 62% |
| RELAXATION MATRIX ANALYSIS OF 2D-NMR DATA | 1990 | 168 | 18 | 78% |
| Study of dynamic processes in liquids using off-resonance rf irradiation | 1999 | 44 | 126 | 33% |
| How to generate accurate solution structures of double-helical nucleic acid fragments using nuclear magnetic resonance and restrained molecular dynamics | 1995 | 70 | 69 | 58% |
| Conformational averaging in structural biology: issues, challenges and computational solutions | 2009 | 16 | 88 | 23% |
| THE 2-DIMENSIONAL TRANSFERRED NUCLEAR OVERHAUSER EFFECT - THEORY AND PRACTICE | 1993 | 104 | 54 | 43% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CNRSUMR C9955INSERMU414 | 1 | 100% | 0.2% | 2 |
| 2 | BIOCHIM THEOR CNRS URA 77 | 1 | 50% | 0.1% | 1 |
| 3 | CEA SERV PHYS ETAT CONDENSE | 1 | 50% | 0.1% | 1 |
| 4 | ERS 133 | 1 | 50% | 0.1% | 1 |
| 5 | INSERM U414 BIOCHIM STRUCT | 1 | 50% | 0.1% | 1 |
| 6 | MOL STRUCTURE | 1 | 50% | 0.1% | 1 |
| 7 | PROT ENGN NETWORK EXECELLENCE | 1 | 50% | 0.1% | 1 |
| 8 | UNITE MIXTE RECH BRUKER | 1 | 50% | 0.1% | 1 |
| 9 | COMMUN RMN | 0 | 10% | 0.4% | 4 |
| 10 | BIO MAT | 0 | 33% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000221157 | SHEARED G CENTER DOT A PAIR//DGGTATACC2//SHEARED A CENTER DOT C PAIR |
| 2 | 0.0000191463 | AFFINITY INDEX//H 1 SPIN LATTICE RELAXATION RATE//LIGAND MACROMOLECULES INTERACTION |
| 3 | 0.0000174599 | JOURNAL OF BIOMOLECULAR NMR//ULTRAFAST 2D NMR//CYANA |
| 4 | 0.0000130701 | CROSS CORRELATED RELAXATION//N 15 RELAXATION//NMR SPIN RELAXATION |
| 5 | 0.0000123833 | GRP BIOPHYS MOL//BIOMOCETIUFR SMBH//UMR 8237 |
| 6 | 0.0000091682 | REDUCTIVE ELIMINATION REACTION//SULFUR INVERSION//TRIMETHYLPLATINUMIV |
| 7 | 0.0000090750 | COMPLEX MOMENTS PROBLEM//RANDOM DETERMINANTS//AUTOMATIC PHASE CORRECTION |
| 8 | 0.0000090193 | R ID STRUCT CHARACTERIZAT GRP//CIGAR HMBC//1 1 ADEQUATE |
| 9 | 0.0000081834 | SOLUTION CONFORMATIONAL ENTROPY//HYDROXY PROTONS//CONFORMATIONAL MAP |
| 10 | 0.0000081442 | DISTANT DIPOLAR FIELD//INTERMOLECULAR MULTIPLE QUANTUM COHERENCES//CRAZED |