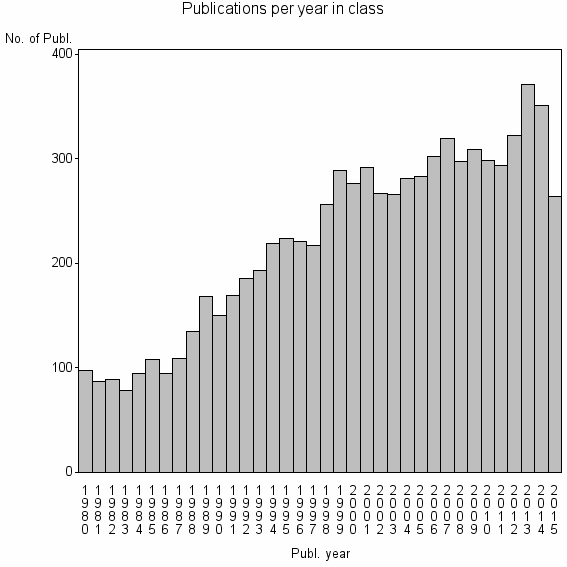
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 1322 | 7974 | 45.3 | 82% |
Classes in level above (level 3) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 136 | 62081 | RNA-A PUBLICATION OF THE RNA SOCIETY//RIBOZYME//NUCLEIC ACIDS RESEARCH |
Classes in level below (level 1) |
| ID, lev. below | Publications | Label for level below |
|---|---|---|
| 4702 | 1666 | IRES//INTERNAL RIBOSOME ENTRY SITE IRES//INTERNAL RIBOSOME ENTRY SITE |
| 5827 | 1500 | EIF3//GENE REGULAT DEV//EIF1 |
| 6073 | 1467 | EIF4E//EIF 4E//CAPPING EFFICIENCY |
| 9295 | 1101 | PKR//VA RNA//NF90 |
| 11891 | 882 | RNA HELICASE//DEAD BOX//DEAD BOX PROTEIN |
| 14481 | 700 | EEF1A2//ELONGATION FACTOR 1 GAMMA//ELONGATION FACTOR 1 |
| 19030 | 458 | ELONGATION FACTOR 2 KINASE//EUKARYOTIC ELONGATION FACTOR 2 EEF2//ELONGATION FACTOR 2 |
| 26849 | 200 | PDCD4//PROGRAMMED CELL DEATH 4//PROGRAMMED CELL DEATH 4 PDCD4 |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | IRES | Author keyword | 154 | 54% | 2% | 196 |
| 2 | TRANSLATION INITIATION | Author keyword | 145 | 40% | 4% | 282 |
| 3 | EIF4E | Author keyword | 143 | 55% | 2% | 179 |
| 4 | PKR | Author keyword | 126 | 51% | 2% | 178 |
| 5 | PDCD4 | Author keyword | 80 | 62% | 1% | 83 |
| 6 | RNA HELICASE | Author keyword | 53 | 39% | 1% | 108 |
| 7 | INITIATION FACTOR | Author keyword | 50 | 61% | 1% | 54 |
| 8 | DEAD BOX | Author keyword | 49 | 66% | 1% | 46 |
| 9 | EIF3 | Author keyword | 48 | 67% | 1% | 43 |
| 10 | EIF4F | Author keyword | 46 | 75% | 0% | 33 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | IRES | 154 | 54% | 2% | 196 | Search IRES | Search IRES |
| 2 | TRANSLATION INITIATION | 145 | 40% | 4% | 282 | Search TRANSLATION+INITIATION | Search TRANSLATION+INITIATION |
| 3 | EIF4E | 143 | 55% | 2% | 179 | Search EIF4E | Search EIF4E |
| 4 | PKR | 126 | 51% | 2% | 178 | Search PKR | Search PKR |
| 5 | PDCD4 | 80 | 62% | 1% | 83 | Search PDCD4 | Search PDCD4 |
| 6 | RNA HELICASE | 53 | 39% | 1% | 108 | Search RNA+HELICASE | Search RNA+HELICASE |
| 7 | INITIATION FACTOR | 50 | 61% | 1% | 54 | Search INITIATION+FACTOR | Search INITIATION+FACTOR |
| 8 | DEAD BOX | 49 | 66% | 1% | 46 | Search DEAD+BOX | Search DEAD+BOX |
| 9 | EIF3 | 48 | 67% | 1% | 43 | Search EIF3 | Search EIF3 |
| 10 | EIF4F | 46 | 75% | 0% | 33 | Search EIF4F | Search EIF4F |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FACTOR EIF 4E | 423 | 90% | 2% | 180 |
| 2 | CAP INDEPENDENT TRANSLATION | 249 | 58% | 4% | 287 |
| 3 | FACTOR 4G EIF4G | 227 | 86% | 1% | 116 |
| 4 | CAP BINDING PROTEIN | 220 | 56% | 3% | 269 |
| 5 | MULTIFACTOR COMPLEX | 206 | 98% | 1% | 52 |
| 6 | INITIATION FACTOR 4E | 201 | 49% | 4% | 303 |
| 7 | TRANSLATION INITIATION | 180 | 24% | 8% | 664 |
| 8 | MESSENGER RNA CAP | 169 | 76% | 1% | 117 |
| 9 | FACTOR 4E | 167 | 71% | 2% | 135 |
| 10 | RIBOSOME ENTRY SITE | 163 | 42% | 4% | 299 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The mechanism of eukaryotic translation initiation and principles of its regulation | 2010 | 570 | 113 | 55% |
| Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets | 2009 | 818 | 128 | 43% |
| The Scanning Mechanism of Eukaryotic Translation Initiation | 2014 | 32 | 228 | 86% |
| A mechanistic overview of translation initiation in eukaryotes | 2012 | 71 | 106 | 96% |
| eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation | 1999 | 1267 | 341 | 65% |
| From unwinding to clamping - the DEAD box RNA helicase family | 2011 | 164 | 137 | 69% |
| The role of IRES trans-acting factors in carcinogenesis | 2015 | 4 | 129 | 57% |
| Translational control in cancer | 2010 | 238 | 139 | 58% |
| Internal ribosome entry sites in eukaryotic mRNA molecules | 2001 | 580 | 170 | 88% |
| Targeting the eukaryotic translation initiation factor 4E for cancer therapy | 2008 | 161 | 19 | 95% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENE REGULAT DEV | 44 | 34% | 1.3% | 105 |
| 2 | MCGILL CANC | 18 | 20% | 1.0% | 79 |
| 3 | TRANSLAT CONTROL GRP | 15 | 77% | 0.1% | 10 |
| 4 | CELLULAR MOL SCI GRP | 13 | 67% | 0.2% | 12 |
| 5 | CNRS URA 1966 | 12 | 86% | 0.1% | 6 |
| 6 | POSTTRANSCRIPT CONTROL GRP | 11 | 57% | 0.2% | 13 |
| 7 | REGULAT GENE EXP S | 9 | 40% | 0.2% | 18 |
| 8 | UNITE REGULAT TRADUCT EUCARYOTE VIRALE | 8 | 70% | 0.1% | 7 |
| 9 | EQUIPE TRADUCT CYCLE CELLULAIRE DEV | 7 | 64% | 0.1% | 7 |
| 10 | EUKARYOT GENE REGULAT | 7 | 21% | 0.4% | 29 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000016636 | PRE MRNA SPLICING//RNA LOCALIZATION//SPLICEOSOME |
| 2 | 0.0000014802 | RIBOSOME//TRANS TRANSLATION//TMRNA |
| 3 | 0.0000012270 | FOOT AND MOUTH DISEASE//ENTEROVIRUS 71//FOOT AND MOUTH DISEASE VIRUS |
| 4 | 0.0000009634 | UNFOLDED PROTEIN RESPONSE//ENDOPLASMIC RETICULUM STRESS//ER STRESS |
| 5 | 0.0000009543 | PTEN//MTOR//LHERMITTE DUCLOS DISEASE |
| 6 | 0.0000008877 | NUCLEOLUS//RIBOSOME BIOGENESIS//NUCLEOLAR ORGANIZER REGIONS |
| 7 | 0.0000008800 | PIM 2//TRANSLATIONALLY CONTROLLED TUMOR PROTEIN//TCTP |
| 8 | 0.0000007933 | PROTEIN KINASE CK2//CASEIN KINASE 2//CK2 |
| 9 | 0.0000007241 | JOURNAL OF INTERFERON RESEARCH//ISG15//JOURNAL OF INTERFERON AND CYTOKINE RESEARCH |
| 10 | 0.0000006772 | RNASE P//AMINOACYL TRNA SYNTHETASE//TRNA |