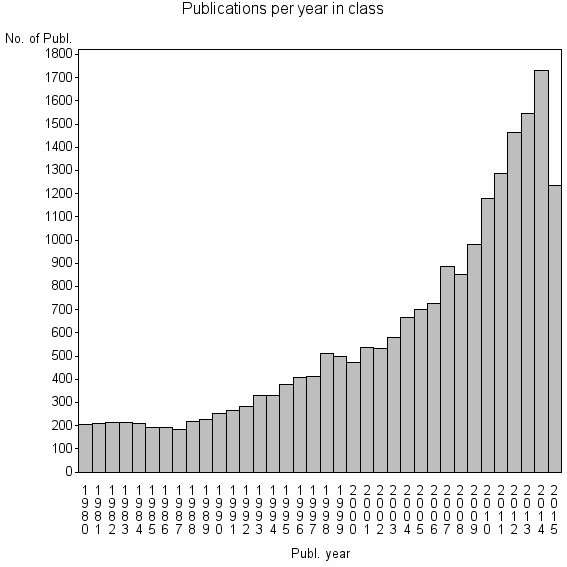
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 181 | 21089 | 49.7 | 87% |
Classes in level above (level 3) |
Classes in level below (level 1) |
| ID, lev. below | Publications | Label for level below |
|---|---|---|
| 380 | 3491 | LINKER HISTONE//HISTONE H1//NUCLEOSOME |
| 1336 | 2614 | HISTONE DEMETHYLASE//HISTONE DEMETHYLATION//LSD1 |
| 2284 | 2226 | EZH2//POLYCOMB//POLYCOMB GROUP |
| 2907 | 2039 | TFIID//TFIIA//MOT1 |
| 4630 | 1680 | CHIP SEQ//PEAK CALLING//DNASE SEQ |
| 4962 | 1628 | P TEFB//CDK9//HEXIM1 |
| 5046 | 1615 | SWI SNF//BRG1//ISWI |
| 5743 | 1515 | HISTONE CHAPERONE//ASF1//CAF 1 |
| 8974 | 1132 | MYST//MOZ//HISTONE ACETYLTRANSFERASE |
| 10294 | 1010 | NUCLEOSOME POSITIONING//NUCLEOSOME//NUCLEOSOME MAPPING |
| 12983 | 804 | MED12//MEDIATOR COMPLEX//MED19 |
| 16638 | 578 | NUCLEAR FACTOR 1//NUCLEAR FACTOR I//DEV GENOM GRP |
| 16900 | 565 | NUT MIDLINE CARCINOMA//BROMODOMAIN//BRD4 |
| 27254 | 192 | REPTIN//PONTIN//R2TP |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CHROMATIN | Author keyword | 345 | 25% | 6% | 1203 |
| 2 | EZH2 | Author keyword | 281 | 65% | 1% | 267 |
| 3 | NUCLEOSOME | Author keyword | 269 | 52% | 2% | 367 |
| 4 | POLYCOMB | Author keyword | 203 | 57% | 1% | 240 |
| 5 | RNA POLYMERASE II | Author keyword | 178 | 41% | 2% | 334 |
| 6 | SWI SNF | Author keyword | 153 | 63% | 1% | 154 |
| 7 | TFIID | Author keyword | 144 | 77% | 0% | 99 |
| 8 | POLYCOMB GROUP | Author keyword | 131 | 75% | 0% | 95 |
| 9 | P TEFB | Author keyword | 127 | 75% | 0% | 92 |
| 10 | BROMODOMAIN | Author keyword | 121 | 71% | 0% | 98 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CHROMATIN | 345 | 25% | 6% | 1203 | Search CHROMATIN | Search CHROMATIN |
| 2 | EZH2 | 281 | 65% | 1% | 267 | Search EZH2 | Search EZH2 |
| 3 | NUCLEOSOME | 269 | 52% | 2% | 367 | Search NUCLEOSOME | Search NUCLEOSOME |
| 4 | POLYCOMB | 203 | 57% | 1% | 240 | Search POLYCOMB | Search POLYCOMB |
| 5 | RNA POLYMERASE II | 178 | 41% | 2% | 334 | Search RNA+POLYMERASE+II | Search RNA+POLYMERASE+II |
| 6 | SWI SNF | 153 | 63% | 1% | 154 | Search SWI+SNF | Search SWI+SNF |
| 7 | TFIID | 144 | 77% | 0% | 99 | Search TFIID | Search TFIID |
| 8 | POLYCOMB GROUP | 131 | 75% | 0% | 95 | Search POLYCOMB+GROUP | Search POLYCOMB+GROUP |
| 9 | P TEFB | 127 | 75% | 0% | 92 | Search P+TEFB | Search P+TEFB |
| 10 | BROMODOMAIN | 121 | 71% | 0% | 98 | Search BROMODOMAIN | Search BROMODOMAIN |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RNA POLYMERASE II | 1185 | 32% | 15% | 3098 |
| 2 | CHROMATIN | 576 | 22% | 11% | 2373 |
| 3 | P TEFB | 412 | 56% | 2% | 507 |
| 4 | H3 | 345 | 50% | 2% | 501 |
| 5 | TATA BINDING PROTEIN | 337 | 39% | 3% | 687 |
| 6 | SWI SNF COMPLEX | 333 | 61% | 2% | 356 |
| 7 | CORE PARTICLE | 323 | 59% | 2% | 363 |
| 8 | NUCLEOSOME | 320 | 47% | 2% | 500 |
| 9 | PREINITIATION COMPLEX | 313 | 57% | 2% | 374 |
| 10 | HISTONE OCTAMER | 297 | 78% | 1% | 197 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR CELL | 54 | 10% | 2% | 503 |
| 2 | EPIGENETICS & CHROMATIN | 24 | 32% | 0% | 64 |
| 3 | BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS | 12 | 12% | 0% | 100 |
| 4 | EPIGENETICS | 11 | 10% | 0% | 103 |
| 5 | METHODS-A COMPANION TO METHODS IN ENZYMOLOGY | 8 | 31% | 0% | 23 |
| 6 | EPIGENOMICS | 8 | 15% | 0% | 46 |
| 7 | NUCLEUS | 1 | 10% | 0% | 14 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Chromatin modifications and their function | 2007 | 3808 | 89 | 56% |
| The Polycomb complex PRC2 and its mark in life | 2011 | 598 | 96 | 80% |
| The role of chromatin during transcription | 2007 | 1530 | 125 | 92% |
| Translating the histone code | 2001 | 4788 | 80 | 39% |
| Histone exchange, chromatin structure and the regulation of transcription | 2015 | 7 | 155 | 93% |
| Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put | 2013 | 144 | 151 | 87% |
| Regulation of chromatin by histone modifications | 2011 | 589 | 121 | 55% |
| Regulation of nucleosome dynamics by histone modifications | 2013 | 159 | 101 | 74% |
| The complex language of chromatin regulation during transcription | 2007 | 1163 | 48 | 79% |
| The Biology of Chromatin Remodeling Complexes | 2009 | 630 | 179 | 78% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NUCLE ACIDS ENZYMOL | 44 | 88% | 0.1% | 21 |
| 2 | RECEPTOR BIOL GENE EXP S | 37 | 32% | 0.5% | 95 |
| 3 | STRUCT GENOM CONSORTIUM | 34 | 21% | 0.7% | 144 |
| 4 | EUKARYOT GENE REGULAT | 29 | 40% | 0.3% | 55 |
| 5 | NUCL ACIDS ENZYMOL | 24 | 82% | 0.1% | 14 |
| 6 | CANC EPIGENET DISCOVERY PERFORMANCE UNIT | 23 | 86% | 0.1% | 12 |
| 7 | GENE REGULAT | 21 | 18% | 0.5% | 110 |
| 8 | HORIKOSHI GENE SELECTOR PROJECT | 21 | 59% | 0.1% | 24 |
| 9 | CHROMATIN BIOL | 19 | 33% | 0.2% | 48 |
| 10 | CHROMATIN GENE EXP S SECT | 19 | 61% | 0.1% | 20 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000016136 | NUCLEAR MATRIX//PROT SECT//MATRIX ATTACHMENT REGION |
| 2 | 0.0000013913 | CHROMOSOMA//SYNAPTONEMAL COMPLEX//MEIOSIS |
| 3 | 0.0000013549 | ARGININE METHYLATION//CARM1//ISOASPARTATE |
| 4 | 0.0000011853 | DNA METHYLATION//EPIGENETICS//HISTONE DEACETYLASE INHIBITOR |
| 5 | 0.0000010093 | XIST//X CHROMOSOME INACTIVATION//TSIX |
| 6 | 0.0000008939 | ASTROCYTE ELEVATED GENE 1//AEG 1//DREF |
| 7 | 0.0000008508 | ID 1//NRSF//MYC |
| 8 | 0.0000008367 | NM23//NUCLEOSIDE DIPHOSPHATE KINASE//ING4 |
| 9 | 0.0000007600 | PRE MRNA SPLICING//RNA LOCALIZATION//SPLICEOSOME |
| 10 | 0.0000007586 | MICROBIAL BIOTECHNOL CELL BIOL//INTERPHASE CHROMATIN//BARLEY CHROMOSOME |