Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
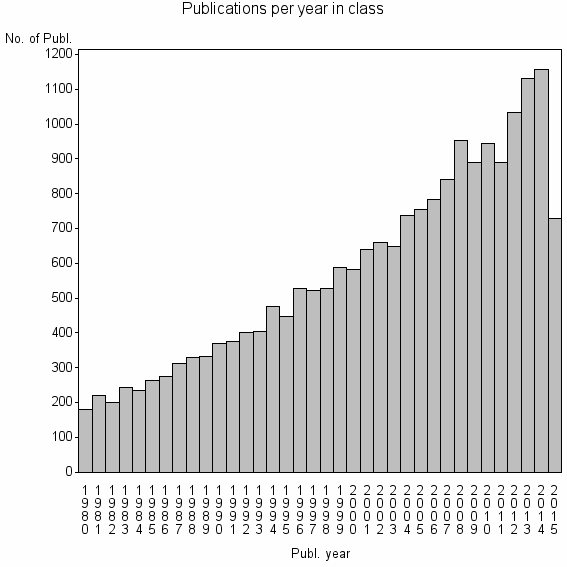
| 195 | 20594 | 47.5 | 85% |
Classes in level above (level 3) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 136 | 62081 | RNA-A PUBLICATION OF THE RNA SOCIETY//RIBOZYME//NUCLEIC ACIDS RESEARCH |
Classes in level below (level 1) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PRE MRNA SPLICING | Author keyword | 275 | 63% | 1% | 277 |
| 2 | RNA LOCALIZATION | Author keyword | 200 | 71% | 1% | 160 |
| 3 | SPLICEOSOME | Author keyword | 198 | 62% | 1% | 205 |
| 4 | RNA BINDING PROTEIN | Author keyword | 172 | 37% | 2% | 372 |
| 5 | RNA-A PUBLICATION OF THE RNA SOCIETY | Journal | 145 | 21% | 3% | 608 |
| 6 | MRNA DECAY | Author keyword | 144 | 59% | 1% | 163 |
| 7 | POLYADENYLATION | Author keyword | 144 | 46% | 1% | 236 |
| 8 | HUR | Author keyword | 138 | 64% | 1% | 134 |
| 9 | SR PROTEINS | Author keyword | 129 | 68% | 1% | 114 |
| 10 | YB 1 | Author keyword | 119 | 71% | 0% | 96 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PRE MRNA SPLICING | 275 | 63% | 1% | 277 | Search PRE+MRNA+SPLICING | Search PRE+MRNA+SPLICING |
| 2 | RNA LOCALIZATION | 200 | 71% | 1% | 160 | Search RNA+LOCALIZATION | Search RNA+LOCALIZATION |
| 3 | SPLICEOSOME | 198 | 62% | 1% | 205 | Search SPLICEOSOME | Search SPLICEOSOME |
| 4 | RNA BINDING PROTEIN | 172 | 37% | 2% | 372 | Search RNA+BINDING+PROTEIN | Search RNA+BINDING+PROTEIN |
| 5 | MRNA DECAY | 144 | 59% | 1% | 163 | Search MRNA+DECAY | Search MRNA+DECAY |
| 6 | POLYADENYLATION | 144 | 46% | 1% | 236 | Search POLYADENYLATION | Search POLYADENYLATION |
| 7 | HUR | 138 | 64% | 1% | 134 | Search HUR | Search HUR |
| 8 | SR PROTEINS | 129 | 68% | 1% | 114 | Search SR+PROTEINS | Search SR+PROTEINS |
| 9 | YB 1 | 119 | 71% | 0% | 96 | Search YB+1 | Search YB+1 |
| 10 | RNA BINDING PROTEINS | 106 | 39% | 1% | 212 | Search RNA+BINDING+PROTEINS | Search RNA+BINDING+PROTEINS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PRE MESSENGER RNA | 1364 | 46% | 11% | 2187 |
| 2 | SR PROTEINS | 638 | 62% | 3% | 652 |
| 3 | AU RICH ELEMENTS | 397 | 58% | 2% | 456 |
| 4 | SMALL NUCLEAR RIBONUCLEOPROTEIN | 354 | 58% | 2% | 407 |
| 5 | EXON JUNCTION COMPLEX | 329 | 66% | 1% | 307 |
| 6 | 3 END FORMATION | 326 | 63% | 2% | 332 |
| 7 | NONSENSE MEDIATED DECAY | 325 | 61% | 2% | 348 |
| 8 | SPLICEOSOME | 312 | 57% | 2% | 370 |
| 9 | DEADENYLATION | 299 | 69% | 1% | 258 |
| 10 | CYTOPLASMIC POLYADENYLATION | 272 | 61% | 1% | 286 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RNA-A PUBLICATION OF THE RNA SOCIETY | 145 | 21% | 3% | 608 |
| 2 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 26 | 27% | 0% | 80 |
| 3 | RNA | 24 | 17% | 1% | 123 |
| 4 | RNA BIOLOGY | 9 | 11% | 0% | 83 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The Spliceosome: Design Principles of a Dynamic RNP Machine | 2009 | 727 | 131 | 76% |
| Expansion of the eukaryotic proteome by alternative splicing | 2010 | 501 | 81 | 51% |
| The nonsense-mediated decay RNA surveillance pathway | 2007 | 539 | 161 | 93% |
| NMD: a multifaceted response to premature translational termination | 2012 | 153 | 179 | 85% |
| Regulation of cytoplasmic mRNA decay | 2012 | 151 | 114 | 84% |
| The highways and byways of mRNA decay | 2007 | 511 | 134 | 83% |
| Mechanisms of alternative pre-messenger RNA splicing | 2003 | 1260 | 352 | 64% |
| The SR protein family of splicing factors: master regulators of gene expression | 2009 | 373 | 206 | 78% |
| P bodies and the control of mRNA translation and degradation | 2007 | 587 | 92 | 70% |
| Alternative mRNA transcription, processing, and translation: insights from RNA sequencing | 2015 | 6 | 139 | 45% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MRNP BIOGENESIS METAB | 42 | 60% | 0.2% | 46 |
| 2 | PROGRAM BIOMOL | 24 | 82% | 0.1% | 14 |
| 3 | HELMHOLTZ JR GRP POSTTRANSCRIPT CONTROL GENE | 19 | 76% | 0.1% | 13 |
| 4 | PROGRAM MOL BIOL BIOTECHNOL | 19 | 22% | 0.4% | 77 |
| 5 | GENOM PHYSIOL | 17 | 75% | 0.1% | 12 |
| 6 | RNA RNP GRP | 17 | 100% | 0.0% | 8 |
| 7 | EDITH JOSEPH FI ER ENZYME INHIBITORS | 15 | 63% | 0.1% | 15 |
| 8 | NEUROEMBRYOL | 13 | 49% | 0.1% | 20 |
| 9 | RNA MOL BIOL | 13 | 18% | 0.3% | 69 |
| 10 | MOL BIOL RNA | 13 | 24% | 0.2% | 47 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000016636 | IRES//TRANSLATION INITIATION//EIF4E |
| 2 | 0.0000011308 | NUCLEOLUS//RIBOSOME BIOGENESIS//NUCLEOLAR ORGANIZER REGIONS |
| 3 | 0.0000009216 | NUCLEAR ENVELOPE//NUCLEAR PORE COMPLEX//NUCLEAR LAMINA |
| 4 | 0.0000007748 | ARGININE METHYLATION//CARM1//ISOASPARTATE |
| 5 | 0.0000007600 | CHROMATIN//EZH2//NUCLEOSOME |
| 6 | 0.0000006787 | PIRNA//RNA INTERFERENCE//PIWI |
| 7 | 0.0000006723 | RNASE P//AMINOACYL TRNA SYNTHETASE//TRNA |
| 8 | 0.0000006668 | ASTROCYTE ELEVATED GENE 1//AEG 1//DREF |
| 9 | 0.0000006258 | NUCLEAR MATRIX//PROT SECT//MATRIX ATTACHMENT REGION |
| 10 | 0.0000006046 | HUMAN ENDOGENOUS RETROVIRUS//ADAR//ENDOGENOUS RETROVIRUS |