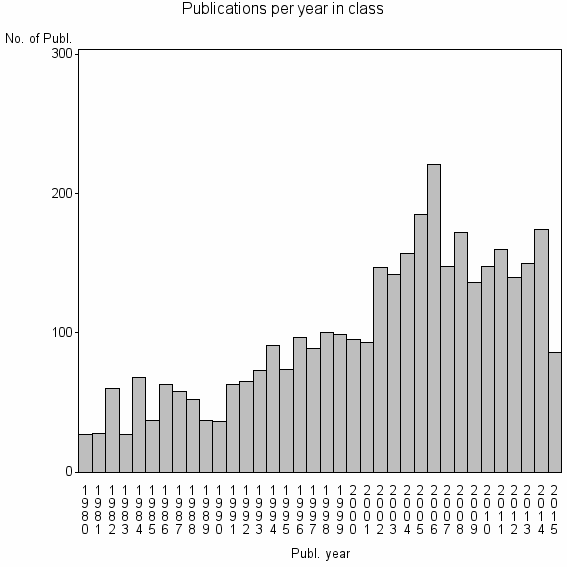
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 2449 | 3598 | 26.3 | 54% |
Classes in level above (level 3) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 596 | 12253 | ORIGINS OF LIFE AND EVOLUTION OF BIOSPHERES//ORIGINS OF LIFE AND EVOLUTION OF THE BIOSPHERE//ORIGIN OF LIFE |
Classes in level below (level 1) |
| ID, lev. below | Publications | Label for level below |
|---|---|---|
| 7461 | 1285 | PREDICTION WITH EXPERT ADVICE//UNIVERSAL CODING//INDIVIDUAL SEQUENCES |
| 10834 | 963 | LATENT PERIODICITY//3 BASE PERIODICITY//DNA WALK |
| 13104 | 796 | GRAPHICAL REPRESENTATION//ALIGNMENT FREE//SIMILARITIES DISSIMILARITIES |
| 20492 | 396 | AMBIGRAM//DESK TOP MOLECULAR MODELING//PARTIAL DIGEST |
| 29062 | 158 | WORD REMOVAL//NORMALIZED COMPRESSION DISTANCE//COMPRESSION BASED DISTANCE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PREDICTION WITH EXPERT ADVICE | Author keyword | 45 | 94% | 0% | 16 |
| 2 | UNIVERSAL CODING | Author keyword | 39 | 53% | 1% | 51 |
| 3 | GRAPHICAL REPRESENTATION | Author keyword | 35 | 38% | 2% | 74 |
| 4 | INDIVIDUAL SEQUENCES | Author keyword | 24 | 68% | 1% | 21 |
| 5 | UNIVERSAL PREDICTION | Author keyword | 22 | 67% | 1% | 20 |
| 6 | MISTAKE BOUNDS | Author keyword | 21 | 85% | 0% | 11 |
| 7 | ALIGNMENT FREE | Author keyword | 20 | 53% | 1% | 27 |
| 8 | UNIVERSAL COMPRESSION | Author keyword | 19 | 80% | 0% | 12 |
| 9 | CHAOS GAME REPRESENTATION | Author keyword | 18 | 53% | 1% | 23 |
| 10 | SIMILARITIES DISSIMILARITIES | Author keyword | 17 | 100% | 0% | 8 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PREDICTION WITH EXPERT ADVICE | 45 | 94% | 0% | 16 | Search PREDICTION+WITH+EXPERT+ADVICE | Search PREDICTION+WITH+EXPERT+ADVICE |
| 2 | UNIVERSAL CODING | 39 | 53% | 1% | 51 | Search UNIVERSAL+CODING | Search UNIVERSAL+CODING |
| 3 | GRAPHICAL REPRESENTATION | 35 | 38% | 2% | 74 | Search GRAPHICAL+REPRESENTATION | Search GRAPHICAL+REPRESENTATION |
| 4 | INDIVIDUAL SEQUENCES | 24 | 68% | 1% | 21 | Search INDIVIDUAL+SEQUENCES | Search INDIVIDUAL+SEQUENCES |
| 5 | UNIVERSAL PREDICTION | 22 | 67% | 1% | 20 | Search UNIVERSAL+PREDICTION | Search UNIVERSAL+PREDICTION |
| 6 | MISTAKE BOUNDS | 21 | 85% | 0% | 11 | Search MISTAKE+BOUNDS | Search MISTAKE+BOUNDS |
| 7 | ALIGNMENT FREE | 20 | 53% | 1% | 27 | Search ALIGNMENT+FREE | Search ALIGNMENT+FREE |
| 8 | UNIVERSAL COMPRESSION | 19 | 80% | 0% | 12 | Search UNIVERSAL+COMPRESSION | Search UNIVERSAL+COMPRESSION |
| 9 | CHAOS GAME REPRESENTATION | 18 | 53% | 1% | 23 | Search CHAOS+GAME+REPRESENTATION | Search CHAOS+GAME+REPRESENTATION |
| 10 | SIMILARITIES DISSIMILARITIES | 17 | 100% | 0% | 8 | Search SIMILARITIES+DISSIMILARITIES | Search SIMILARITIES+DISSIMILARITIES |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SIMILARITY DISSIMILARITY | 276 | 94% | 3% | 97 |
| 2 | NUMERICAL CHARACTERIZATION | 266 | 74% | 6% | 198 |
| 3 | 2 D GRAPHICAL REPRESENTATION | 259 | 82% | 4% | 151 |
| 4 | H CURVES | 102 | 100% | 1% | 30 |
| 5 | CHAOS GAME REPRESENTATION | 88 | 65% | 2% | 83 |
| 6 | DNA PRIMARY SEQUENCES | 87 | 72% | 2% | 68 |
| 7 | DUAL NUCLEOTIDES | 79 | 88% | 1% | 37 |
| 8 | 1 F ALPHA SPECTRUM | 61 | 95% | 1% | 20 |
| 9 | LOW DEGENERACY | 60 | 100% | 1% | 20 |
| 10 | INDIVIDUAL SEQUENCES | 42 | 44% | 2% | 71 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Graphical Representation of Proteins | 2011 | 46 | 162 | 53% |
| Alignment-free sequence comparison - a review | 2003 | 279 | 47 | 38% |
| Proteomics, networks and connectivity indices | 2008 | 157 | 259 | 21% |
| Milestones in Graphical Bioinformatics | 2013 | 8 | 117 | 63% |
| Gene Prediction Based on DNA Spectral Analysis: A Literature Review | 2011 | 15 | 34 | 71% |
| Universal prediction | 1998 | 129 | 50 | 82% |
| Model selection and the principle of minimum description length | 2001 | 233 | 44 | 39% |
| On a 3-D representation of DNA primary sequences | 2004 | 34 | 10 | 100% |
| The study of correlation structures of DNA sequences: a critical review | 1997 | 128 | 79 | 52% |
| Online Learning with (Multiple) Kernels: A Review | 2013 | 5 | 35 | 37% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GRP INFORMAT ELECT | 15 | 82% | 0.3% | 9 |
| 2 | T LIFE | 6 | 30% | 0.5% | 18 |
| 3 | COMP LEARNING | 6 | 25% | 0.5% | 19 |
| 4 | PROGRAM STAT OPERAT | 5 | 44% | 0.2% | 8 |
| 5 | COMP PRINCIPLES | 4 | 75% | 0.1% | 3 |
| 6 | A YA USIKOV RADIOPHYS ELECT | 3 | 17% | 0.5% | 19 |
| 7 | PHYS BIOL UNIT | 3 | 50% | 0.1% | 4 |
| 8 | BETH ISRAEL HOSP MED CARDIOVASC | 3 | 60% | 0.1% | 3 |
| 9 | CIELO | 2 | 67% | 0.1% | 2 |
| 10 | COMP LEARNING ROYAL HOLLOWAY | 2 | 67% | 0.1% | 2 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000014892 | VALENCE TOPOLOGICAL CHARGE TRANSFER INDEX//UNIV CIENCIA MOL//STELLA AVRAM GOREN GOLDSTEIN BIOTECHNOL EN |
| 2 | 0.0000013580 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
| 3 | 0.0000011205 | STRAY VOLTAGE//COMPLEMENTARY PEPTIDES//COMPLEMENTARY PEPTIDE |
| 4 | 0.0000009745 | HOMOTOPIC MAPPING//CORRELATION DIMENSION//SURROGATE DATA |
| 5 | 0.0000008361 | IEEE TRANSACTIONS ON CIRCUITS AND SYSTEMS FOR VIDEO TECHNOLOGY//SIGNAL PROCESSING-IMAGE COMMUNICATION//VIDEO CODING |
| 6 | 0.0000007112 | HEART RATE VARIABILITY//RESPIRATORY SINUS ARRHYTHMIA//AUTONOMIC NERVOUS SYSTEM |
| 7 | 0.0000006763 | RIBOSOME//TRANS TRANSLATION//TMRNA |
| 8 | 0.0000006556 | APPARENT EQUILIBRIUM CONSTANTS//TRANSFORMED THERMODYNAMIC PROPERTIES//TRANSFORMED GIBBS ENERGY |
| 9 | 0.0000006087 | CREDIT SCORING//MACHINE LEARNING//FEATURE SELECTION |
| 10 | 0.0000005919 | DIRICHLET PROCESS//MARKOV CHAIN MONTE CARLO//BAYESIAN NONPARAMETRICS |