Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
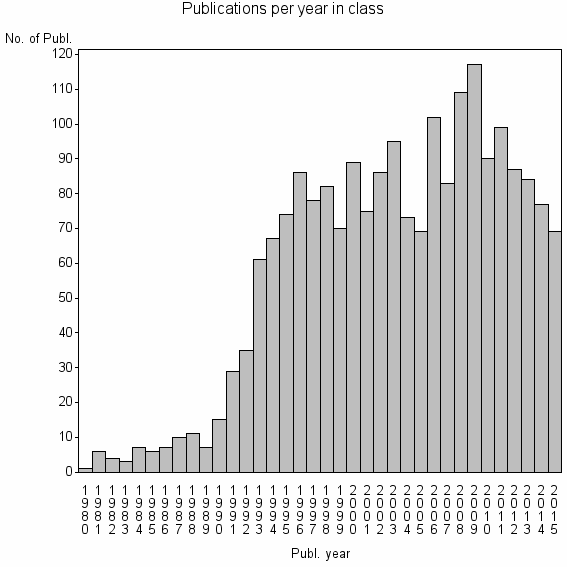
| 3013 | 2063 | 40.7 | 87% |
Classes in level above (level 3) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 465 | 22192 | NM23//RIBONUCLEOTIDE REDUCTASE//DEOXYCYTIDINE KINASE |
Classes in level below (level 1) |
| ID, lev. below | Publications | Label for level below |
|---|---|---|
| 6803 | 1365 | NM23//NUCLEOSIDE DIPHOSPHATE KINASE//NM23 H1 |
| 20430 | 399 | ING4//ING1//P33ING1 |
| 23206 | 299 | BRMS1//BREAST CANCER METASTASIS SUPPRESSOR 1//FDN CANC METASTASIS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | NM23 | Author keyword | 405 | 81% | 12% | 245 |
| 2 | NUCLEOSIDE DIPHOSPHATE KINASE | Author keyword | 187 | 67% | 8% | 167 |
| 3 | ING4 | Author keyword | 118 | 95% | 2% | 39 |
| 4 | NM23 H1 | Author keyword | 116 | 74% | 4% | 87 |
| 5 | ING1 | Author keyword | 113 | 91% | 2% | 48 |
| 6 | NDP KINASE | Author keyword | 82 | 86% | 2% | 42 |
| 7 | BRMS1 | Author keyword | 62 | 80% | 2% | 39 |
| 8 | NDPK | Author keyword | 50 | 77% | 2% | 34 |
| 9 | P33ING1 | Author keyword | 42 | 94% | 1% | 15 |
| 10 | P33ING1B | Author keyword | 41 | 100% | 1% | 15 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NM23 | 405 | 81% | 12% | 245 | Search NM23 | Search NM23 |
| 2 | NUCLEOSIDE DIPHOSPHATE KINASE | 187 | 67% | 8% | 167 | Search NUCLEOSIDE+DIPHOSPHATE+KINASE | Search NUCLEOSIDE+DIPHOSPHATE+KINASE |
| 3 | ING4 | 118 | 95% | 2% | 39 | Search ING4 | Search ING4 |
| 4 | NM23 H1 | 116 | 74% | 4% | 87 | Search NM23+H1 | Search NM23+H1 |
| 5 | ING1 | 113 | 91% | 2% | 48 | Search ING1 | Search ING1 |
| 6 | NDP KINASE | 82 | 86% | 2% | 42 | Search NDP+KINASE | Search NDP+KINASE |
| 7 | BRMS1 | 62 | 80% | 2% | 39 | Search BRMS1 | Search BRMS1 |
| 8 | NDPK | 50 | 77% | 2% | 34 | Search NDPK | Search NDPK |
| 9 | P33ING1 | 42 | 94% | 1% | 15 | Search P33ING1 | Search P33ING1 |
| 10 | P33ING1B | 41 | 100% | 1% | 15 | Search P33ING1B | Search P33ING1B |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NUCLEOSIDE DIPHOSPHATE KINASE | 403 | 54% | 25% | 520 |
| 2 | P33ING1 | 147 | 94% | 2% | 51 |
| 3 | NM23 | 112 | 70% | 5% | 93 |
| 4 | DUCTAL BREAST CARCINOMAS | 98 | 82% | 3% | 58 |
| 5 | GOOD PROGNOSIS | 82 | 65% | 4% | 78 |
| 6 | TUMOR METASTASIS | 69 | 18% | 16% | 339 |
| 7 | DIPHOSPHOKINASE | 58 | 92% | 1% | 23 |
| 8 | NM23 GENE | 58 | 80% | 2% | 36 |
| 9 | PHD FINGER PROTEINS | 56 | 100% | 1% | 19 |
| 10 | DROSOPHILA DEVELOPMENT | 54 | 39% | 5% | 108 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The ING tumor suppressor genes: Status in human tumors | 2014 | 6 | 155 | 90% |
| Nucleoside diphosphate kinase as protein histidine kinase | 2015 | 1 | 48 | 58% |
| The mammalian Nm23/NDPK family: from metastasis control to cilia movement | 2009 | 56 | 102 | 75% |
| The new tumor suppressor genes ING: Genomic structure and status in cancer | 2008 | 50 | 57 | 98% |
| Keep-ING balance: Tumor suppression by epigenetic regulation | 2014 | 5 | 205 | 70% |
| ING1 and ING2: multifaceted tumor suppressor genes | 2013 | 12 | 127 | 72% |
| The ING Gene Family in the Regulation of Cell Growth and Tumorigenesis | 2009 | 50 | 133 | 77% |
| Phosphorylation of basic amino acid residues in proteins: important but easily missed | 2011 | 41 | 128 | 51% |
| Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene | 2012 | 13 | 118 | 88% |
| Different HATS of the ING1 gene family | 2002 | 121 | 55 | 75% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | WOMENS CANC SECT | 14 | 39% | 1.4% | 28 |
| 2 | BIOL DIFFERENCIAT DEV | 11 | 67% | 0.5% | 10 |
| 3 | FDN CANC METASTASIS | 11 | 78% | 0.3% | 7 |
| 4 | UNITE REGULAT ENZYMAT ACT CELLULAI | 6 | 41% | 0.5% | 11 |
| 5 | EA 3674 | 3 | 60% | 0.1% | 3 |
| 6 | UNITE REGULAT ENZYMAT ACTIV CELLULAI | 2 | 38% | 0.2% | 5 |
| 7 | ANAT CYTODIAGNOST PATHOL UNIT | 2 | 67% | 0.1% | 2 |
| 8 | BIOCHEM MOL BIOL ONCOL | 2 | 67% | 0.1% | 2 |
| 9 | CVS LUNG BIOL | 2 | 67% | 0.1% | 2 |
| 10 | EA DRED 483 | 2 | 67% | 0.1% | 2 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000026165 | ADENYLATE KINASE//THYMIDYLATE KINASE//CHIM STRUCT MACROMOL |
| 2 | 0.0000014313 | CHROMATIN SUPRAORGANIZATION//ZNF703//NUCLEAR PHENOTYPES |
| 3 | 0.0000010686 | TETRASPANIN//CD9//CD151 |
| 4 | 0.0000009507 | MASPIN//RARRES3//TIG3 |
| 5 | 0.0000008367 | CHROMATIN//EZH2//NUCLEOSOME |
| 6 | 0.0000007766 | RKIP//RAF KINASE INHIBITOR PROTEIN//HIPPOCAMPAL CHOLINERGIC NEUROSTIMULATING PEPTIDE |
| 7 | 0.0000007159 | N PHOSPHORYL AMINO ACID//BIOORGAN PHOSPHORUS CHEM//SERYL HISTIDINE DIPEPTIDE |
| 8 | 0.0000006768 | EPITHELIAL MESENCHYMAL TRANSITION//EMT//SNAIL |
| 9 | 0.0000006343 | ID 1//NRSF//MYC |
| 10 | 0.0000005721 | 3 ISOPROPYLMALATE DEHYDROGENASE//GLUTAMATE DEHYDROGENASE//ISOCITRATE DEHYDROGENASE |