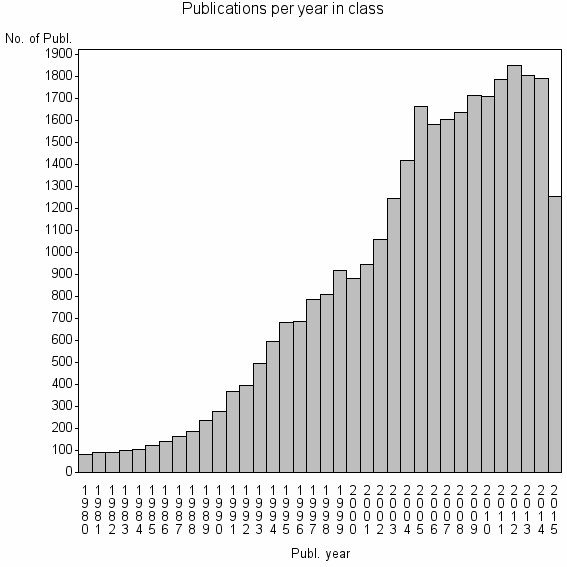
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 42 | 31225 | 44.9 | 79% |
Classes in level above (level 3) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 204 | 48455 | PROTEIN FOLDING//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION |
Classes in level below (level 1) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | Journal | 936 | 35% | 7% | 2147 |
| 2 | PROTEIN STRUCTURE PREDICTION | Author keyword | 677 | 68% | 2% | 600 |
| 3 | PROTEIN FOLDING | Author keyword | 563 | 29% | 5% | 1651 |
| 4 | PSEUDO AMINO ACID COMPOSITION | Author keyword | 329 | 94% | 0% | 119 |
| 5 | FOLD RECOGNITION | Author keyword | 185 | 68% | 1% | 161 |
| 6 | PROTEIN SCIENCE | Journal | 142 | 15% | 3% | 884 |
| 7 | CASP | Author keyword | 135 | 74% | 0% | 100 |
| 8 | STRUCTURE PREDICTION | Author keyword | 129 | 39% | 1% | 265 |
| 9 | COMPUTATIONAL PROTEIN DESIGN | Author keyword | 129 | 79% | 0% | 84 |
| 10 | BETA HAIRPIN | Author keyword | 123 | 66% | 0% | 114 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN STRUCTURE PREDICTION | 677 | 68% | 2% | 600 | Search PROTEIN+STRUCTURE+PREDICTION | Search PROTEIN+STRUCTURE+PREDICTION |
| 2 | PROTEIN FOLDING | 563 | 29% | 5% | 1651 | Search PROTEIN+FOLDING | Search PROTEIN+FOLDING |
| 3 | PSEUDO AMINO ACID COMPOSITION | 329 | 94% | 0% | 119 | Search PSEUDO+AMINO+ACID+COMPOSITION | Search PSEUDO+AMINO+ACID+COMPOSITION |
| 4 | FOLD RECOGNITION | 185 | 68% | 1% | 161 | Search FOLD+RECOGNITION | Search FOLD+RECOGNITION |
| 5 | CASP | 135 | 74% | 0% | 100 | Search CASP | Search CASP |
| 6 | STRUCTURE PREDICTION | 129 | 39% | 1% | 265 | Search STRUCTURE+PREDICTION | Search STRUCTURE+PREDICTION |
| 7 | COMPUTATIONAL PROTEIN DESIGN | 129 | 79% | 0% | 84 | Search COMPUTATIONAL+PROTEIN+DESIGN | Search COMPUTATIONAL+PROTEIN+DESIGN |
| 8 | BETA HAIRPIN | 123 | 66% | 0% | 114 | Search BETA+HAIRPIN | Search BETA+HAIRPIN |
| 9 | THREADING | 108 | 56% | 0% | 132 | Search THREADING | Search THREADING |
| 10 | PROTEIN STRUCTURE | 96 | 16% | 2% | 567 | Search PROTEIN+STRUCTURE | Search PROTEIN+STRUCTURE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GLOBULAR PROTEINS | 815 | 38% | 5% | 1689 |
| 2 | FOLD RECOGNITION | 619 | 70% | 2% | 513 |
| 3 | FUNCTIONAL DOMAIN COMPOSITION | 498 | 89% | 1% | 221 |
| 4 | CONTACT ORDER | 495 | 82% | 1% | 291 |
| 5 | CHYMOTRYPSIN INHIBITOR 2 | 491 | 75% | 1% | 356 |
| 6 | STRUCTURE PREDICTION | 426 | 33% | 3% | 1075 |
| 7 | ENZYME SUBFAMILY CLASSES | 422 | 97% | 0% | 115 |
| 8 | SUBCELLULAR LOCATION PREDICTION | 420 | 91% | 1% | 171 |
| 9 | AMINO ACID COMPOSITION | 420 | 40% | 3% | 834 |
| 10 | STRUCTURAL CLASS PREDICTION | 414 | 91% | 1% | 169 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 936 | 35% | 7% | 2147 |
| 2 | PROTEIN SCIENCE | 142 | 15% | 3% | 884 |
| 3 | BIOINFORMATICS | 121 | 11% | 3% | 1048 |
| 4 | JOURNAL OF COMPUTATIONAL CHEMISTRY | 121 | 14% | 3% | 803 |
| 5 | JOURNAL OF CHEMICAL THEORY AND COMPUTATION | 105 | 16% | 2% | 609 |
| 6 | PROTEIN ENGINEERING | 87 | 19% | 1% | 419 |
| 7 | BMC BIOINFORMATICS | 73 | 10% | 2% | 674 |
| 8 | FOLDING & DESIGN | 66 | 51% | 0% | 91 |
| 9 | BMC STRUCTURAL BIOLOGY | 57 | 32% | 0% | 151 |
| 10 | CURRENT OPINION IN STRUCTURAL BIOLOGY | 49 | 15% | 1% | 310 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| CHARMM: The Biomolecular Simulation Program | 2009 | 2048 | 616 | 44% |
| Intrinsically disordered proteins in cellular signalling and regulation | 2015 | 17 | 138 | 53% |
| The Amber biomolecular simulation programs | 2005 | 2779 | 124 | 52% |
| Some remarks on protein attribute prediction and pseudo amino acid composition | 2011 | 329 | 197 | 90% |
| iRSpot-TNCPseAAC: Identify Recombination Spots with Trinucleotide Composition and Pseudo Amino Acid Components | 2014 | 48 | 110 | 88% |
| Locating proteins in the cell using TargetP, SignalP and related tools | 2007 | 1418 | 110 | 41% |
| All-atom empirical potential for molecular modeling and dynamics studies of proteins | 1998 | 6458 | 78 | 32% |
| Interfaces and the driving force of hydrophobic assembly | 2005 | 1314 | 46 | 70% |
| The ensemble nature of allostery | 2014 | 69 | 91 | 47% |
| Recent progress in protein subcellular location prediction | 2007 | 474 | 77 | 91% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | COMPUTAT BIOL BIOINFORMAT | 106 | 27% | 1.1% | 345 |
| 2 | BAKER CHEM CHEM BIOL | 105 | 51% | 0.5% | 145 |
| 3 | SACKLER MOL MED | 98 | 47% | 0.5% | 152 |
| 4 | COMPUTAT MUTAT PROJECT | 82 | 92% | 0.1% | 33 |
| 5 | CUBIC | 60 | 72% | 0.2% | 48 |
| 6 | CHIM BIOPHYS | 59 | 45% | 0.3% | 98 |
| 7 | BIOL RUMENTAT | 54 | 34% | 0.4% | 130 |
| 8 | STUDY SYST BIOL | 50 | 64% | 0.2% | 49 |
| 9 | THEORET BIOL PHYS | 48 | 23% | 0.6% | 180 |
| 10 | BIOMOL STRUCT MODELLING UNIT | 47 | 68% | 0.1% | 41 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000017352 | MOLTEN GLOBULE//APOMYOGLOBIN//PROTEIN FORMULATION |
| 2 | 0.0000014465 | DISSIPATIVE PARTICLE DYNAMICS//FLUCTUATING HYDRODYNAMICS//MADELUNG CONSTANT |
| 3 | 0.0000013077 | CELL FREE PROTEIN SYNTHESIS//MEMBRANE PROTEIN//RIBOSOME DISPLAY |
| 4 | 0.0000011671 | VALENCE TOPOLOGICAL CHARGE TRANSFER INDEX//UNIV CIENCIA MOL//STELLA AVRAM GOREN GOLDSTEIN BIOTECHNOL EN |
| 5 | 0.0000011561 | JOURNAL OF CHEMICAL INFORMATION AND MODELING//B IT//JOURNAL OF CHEMICAL INFORMATION AND COMPUTER SCIENCES |
| 6 | 0.0000011325 | UNITA RIC ROMA TRE//DYNAMICAL TRANSITION//UFR PHITEM |
| 7 | 0.0000010947 | VALLEY RIDGE INFLECTION POINT//NEWTON TRAJECTORY//GRADIENT EXTREMAL |
| 8 | 0.0000010627 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
| 9 | 0.0000010624 | ARTIFICIAL METALLOENZYMES//COILED COIL//PROTEIN DESIGN |
| 10 | 0.0000010559 | HIV 1 PROTEASE//HIV PROTEASE//KNI 272 |