Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
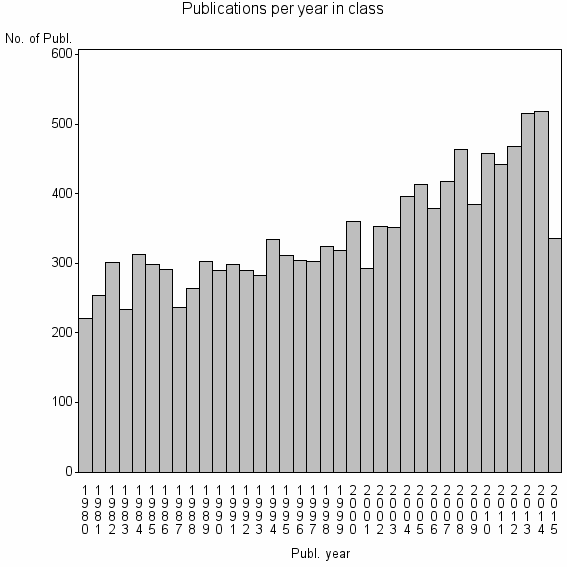
| 695 | 12311 | 44.7 | 71% |
Classes in level above (level 3) |
Classes in level below (level 1) |
| ID, lev. below | Publications | Label for level below |
|---|---|---|
| 1164 | 2710 | SYNAPTONEMAL COMPLEX//MEIOSIS//SYNAPSIS |
| 3121 | 1978 | P ELEMENT//HYBRID DYSGENESIS//HOBO ELEMENT |
| 3706 | 1848 | CHROMOSOME TERRITORY//CHROMOSOME TERRITORIES//NUCLEAR ARCHITECTURE |
| 7737 | 1255 | HP1//POSITION EFFECT VARIEGATION//HETEROCHROMATIN |
| 9193 | 1110 | POLYTENE CHROMOSOMES//CHIRONOMUS//BALBIANI RING GENES |
| 9436 | 1087 | SEX LETHAL//MSL COMPLEX//DOSAGE COMPENSATION |
| 11489 | 911 | CTCF//CONTROL GENET PROC//ENHANCER BLOCKING |
| 17566 | 529 | POKEY//LAMPBRUSH LOOPS//FERTILITY GENES |
| 22630 | 319 | DNA PUFF//DNA PUFFS//RHYNCHOSCIARA |
| 27200 | 193 | CHROMATIN DIMINUTION//GERM LINE LIMITED CHROMOSOMES//PARASCARIS UNIVALENS |
| 29523 | 151 | ABNORMAL MITOSES//CELL POPULAT GENET//MORPHOSIS |
| 31874 | 117 | ASCOBOLUS//EVOLUTION OF PLOIDY//MULTIGENE ARRAYS |
| 32755 | 103 | CALIBRACHOA//PETUNIA INTEGRIFOLIA//PETUNIA |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CHROMOSOMA | Journal | 217 | 27% | 6% | 695 |
| 2 | SYNAPTONEMAL COMPLEX | Author keyword | 141 | 51% | 2% | 196 |
| 3 | MEIOSIS | Author keyword | 120 | 18% | 5% | 624 |
| 4 | CONTROL GENET PROC | Address | 99 | 88% | 0% | 46 |
| 5 | CHROMOSOME TERRITORY | Author keyword | 93 | 81% | 0% | 56 |
| 6 | P ELEMENT | Author keyword | 68 | 59% | 1% | 77 |
| 7 | HETEROCHROMATIN | Author keyword | 67 | 20% | 2% | 297 |
| 8 | CHROMOSOME TERRITORIES | Author keyword | 61 | 77% | 0% | 41 |
| 9 | CTCF | Author keyword | 59 | 51% | 1% | 83 |
| 10 | NUCLEAR ARCHITECTURE | Author keyword | 58 | 47% | 1% | 91 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SYNAPTONEMAL COMPLEX | 141 | 51% | 2% | 196 | Search SYNAPTONEMAL+COMPLEX | Search SYNAPTONEMAL+COMPLEX |
| 2 | MEIOSIS | 120 | 18% | 5% | 624 | Search MEIOSIS | Search MEIOSIS |
| 3 | CHROMOSOME TERRITORY | 93 | 81% | 0% | 56 | Search CHROMOSOME+TERRITORY | Search CHROMOSOME+TERRITORY |
| 4 | P ELEMENT | 68 | 59% | 1% | 77 | Search P+ELEMENT | Search P+ELEMENT |
| 5 | HETEROCHROMATIN | 67 | 20% | 2% | 297 | Search HETEROCHROMATIN | Search HETEROCHROMATIN |
| 6 | CHROMOSOME TERRITORIES | 61 | 77% | 0% | 41 | Search CHROMOSOME+TERRITORIES | Search CHROMOSOME+TERRITORIES |
| 7 | CTCF | 59 | 51% | 1% | 83 | Search CTCF | Search CTCF |
| 8 | NUCLEAR ARCHITECTURE | 58 | 47% | 1% | 91 | Search NUCLEAR+ARCHITECTURE | Search NUCLEAR+ARCHITECTURE |
| 9 | SEX LETHAL | 58 | 75% | 0% | 42 | Search SEX+LETHAL | Search SEX+LETHAL |
| 10 | POSITION EFFECT VARIEGATION | 52 | 61% | 0% | 55 | Search POSITION+EFFECT+VARIEGATION | Search POSITION+EFFECT+VARIEGATION |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CHROMOSOME SYNAPSIS | 409 | 64% | 3% | 404 |
| 2 | SYNAPTONEMAL COMPLEX FORMATION | 399 | 83% | 2% | 228 |
| 3 | SYNAPTONEMAL COMPLEX | 350 | 60% | 3% | 385 |
| 4 | POSITION EFFECT VARIEGATION | 345 | 49% | 4% | 517 |
| 5 | CHIASMA FORMATION | 233 | 88% | 1% | 112 |
| 6 | CROSSING OVER | 228 | 49% | 3% | 342 |
| 7 | MEIOSIS | 225 | 25% | 6% | 794 |
| 8 | CHROMOSOME TERRITORIES | 204 | 58% | 2% | 235 |
| 9 | ORDER CHROMATIN ARRANGEMENTS | 182 | 91% | 1% | 77 |
| 10 | MEIOTIC CHROMOSOME SYNAPSIS | 181 | 76% | 1% | 128 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CHROMOSOMA | 217 | 27% | 6% | 695 |
| 2 | CHROMOSOME RESEARCH | 22 | 11% | 1% | 182 |
| 3 | GENETICAL RESEARCH | 19 | 14% | 1% | 126 |
| 4 | NUCLEUS-AUSTIN | 3 | 12% | 0% | 26 |
| 5 | EPIGENETICS & CHROMATIN | 3 | 11% | 0% | 22 |
| 6 | NUCLEUS | 2 | 12% | 0% | 16 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| CTCF: an architectural protein bridging genome topology and function | 2014 | 56 | 99 | 51% |
| Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data | 2013 | 116 | 105 | 71% |
| CTCF: Master Weaver of the Genome | 2009 | 538 | 99 | 55% |
| The Role of Chromosome Domains in Shaping the Functional Genome | 2015 | 8 | 95 | 61% |
| A decade of 3C technologies: insights into nuclear organization | 2012 | 166 | 96 | 74% |
| The Hierarchy of the 3D Genome | 2013 | 107 | 100 | 61% |
| Nuclear organization of the genome and the potential for gene regulation | 2007 | 380 | 41 | 78% |
| Heterochromatin revisited | 2007 | 524 | 140 | 58% |
| Beyond the sequence: Cellular organization of genome function | 2007 | 501 | 114 | 56% |
| Genome Architecture: Domain Organization of Interphase Chromosomes | 2013 | 95 | 118 | 61% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CONTROL GENET PROC | 99 | 88% | 0.4% | 46 |
| 2 | GENE BIOL | 24 | 16% | 1.1% | 136 |
| 3 | PROLIFERAC CELULAR DESARROLLO | 21 | 90% | 0.1% | 9 |
| 4 | GENOME FUNCT GRP | 15 | 88% | 0.1% | 7 |
| 5 | OPT MICROSCOPY | 13 | 71% | 0.1% | 10 |
| 6 | NUCL DYNAM PROGRAMME | 12 | 75% | 0.1% | 9 |
| 7 | DYNAM GENOME EVOLUT | 11 | 40% | 0.2% | 21 |
| 8 | IMMUNOEPIGENET | 10 | 73% | 0.1% | 8 |
| 9 | CHROMOSOME BIOL | 10 | 20% | 0.4% | 44 |
| 10 | GRP TRANSCRIPT REGULAT | 9 | 83% | 0.0% | 5 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000019068 | XIST//X CHROMOSOME INACTIVATION//TSIX |
| 2 | 0.0000015433 | MICROBIAL BIOTECHNOL CELL BIOL//INTERPHASE CHROMATIN//BARLEY CHROMOSOME |
| 3 | 0.0000013913 | CHROMATIN//EZH2//NUCLEOSOME |
| 4 | 0.0000011837 | KINETOCHORE//MITOSIS//CENTROSOME |
| 5 | 0.0000011272 | DIOECIOUS PLANT//PLANT DEV GENET//SILENE LATIFOLIA |
| 6 | 0.0000010546 | NUCLEAR ENVELOPE//NUCLEAR PORE COMPLEX//NUCLEAR LAMINA |
| 7 | 0.0000010186 | HUMAN ENDOGENOUS RETROVIRUS//ADAR//ENDOGENOUS RETROVIRUS |
| 8 | 0.0000009615 | MADS BOX GENES//MADS BOX//FLOWER DEVELOPMENT |
| 9 | 0.0000009531 | HOLOKINETIC CHROMOSOMES//PHASMATODEA//SEX DETERMINING SYSTEM |
| 10 | 0.0000009282 | NUCLEAR MATRIX//PROT SECT//MATRIX ATTACHMENT REGION |