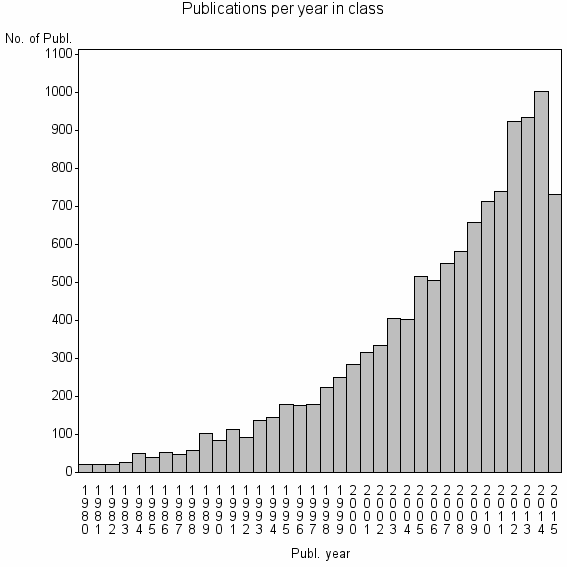
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 777 | 11593 | 50.6 | 85% |
Classes in level above (level 3) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 4 | 177718 | PLANT SCIENCES//PLANT PHYSIOLOGY//PLANT CELL REPORTS |
Classes in level below (level 1) |
| ID, lev. below | Publications | Label for level below |
|---|---|---|
| 2501 | 2153 | MSAP//RNA DIRECTED DNA METHYLATION//RDDM |
| 3110 | 1981 | MADS BOX GENES//MADS BOX//FLOWER DEVELOPMENT |
| 3202 | 1962 | MIR156//DCL1//MIR172 |
| 3821 | 1827 | FLOWERING TIME//FLORIGEN//FLOWERING LOCUS T |
| 5143 | 1597 | RETROTRANSPOSONS//TY1 COPIA//TRANSPOSABLE ELEMENTS |
| 6481 | 1410 | AC DS//TRANSPOSON TAGGING//AC TRANSPOSASE |
| 21755 | 349 | RICE GENOME PROGRAM//CROP DESIGN//GENOME IL |
| 27010 | 197 | PLANT MICROBIAL GENOM//LRT1//NONADDITIVE PROTEIN |
| 31868 | 117 | BRACHYPODIUM DISTACHYON//BRACHYPODIUM//BRACHYPODIUM HYBRIDUM |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MADS BOX GENES | Author keyword | 453 | 86% | 2% | 233 |
| 2 | MADS BOX | Author keyword | 213 | 70% | 2% | 179 |
| 3 | FLOWER DEVELOPMENT | Author keyword | 173 | 49% | 2% | 258 |
| 4 | AGAMOUS | Author keyword | 146 | 88% | 1% | 70 |
| 5 | FLOWERING TIME | Author keyword | 140 | 41% | 2% | 268 |
| 6 | APETALA3 | Author keyword | 106 | 91% | 0% | 43 |
| 7 | MIR156 | Author keyword | 98 | 94% | 0% | 34 |
| 8 | ABC MODEL | Author keyword | 89 | 74% | 1% | 66 |
| 9 | MSAP | Author keyword | 81 | 71% | 1% | 65 |
| 10 | FLORAL ORGAN IDENTITY | Author keyword | 77 | 89% | 0% | 34 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MADS BOX GENES | 453 | 86% | 2% | 233 | Search MADS+BOX+GENES | Search MADS+BOX+GENES |
| 2 | MADS BOX | 213 | 70% | 2% | 179 | Search MADS+BOX | Search MADS+BOX |
| 3 | FLOWER DEVELOPMENT | 173 | 49% | 2% | 258 | Search FLOWER+DEVELOPMENT | Search FLOWER+DEVELOPMENT |
| 4 | AGAMOUS | 146 | 88% | 1% | 70 | Search AGAMOUS | Search AGAMOUS |
| 5 | FLOWERING TIME | 140 | 41% | 2% | 268 | Search FLOWERING+TIME | Search FLOWERING+TIME |
| 6 | APETALA3 | 106 | 91% | 0% | 43 | Search APETALA3 | Search APETALA3 |
| 7 | MIR156 | 98 | 94% | 0% | 34 | Search MIR156 | Search MIR156 |
| 8 | ABC MODEL | 89 | 74% | 1% | 66 | Search ABC+MODEL | Search ABC+MODEL |
| 9 | MSAP | 81 | 71% | 1% | 65 | Search MSAP | Search MSAP |
| 10 | FLORAL ORGAN IDENTITY | 77 | 89% | 0% | 34 | Search FLORAL+ORGAN+IDENTITY | Search FLORAL+ORGAN+IDENTITY |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PLANT MICRORNAS | 828 | 85% | 4% | 433 |
| 2 | FLOWER DEVELOPMENT | 809 | 58% | 8% | 940 |
| 3 | MADS BOX GENES | 806 | 62% | 7% | 838 |
| 4 | FLORAL ORGAN IDENTITY | 607 | 86% | 3% | 310 |
| 5 | ORGAN IDENTITY | 561 | 90% | 2% | 246 |
| 6 | MERISTEM IDENTITY | 448 | 78% | 3% | 299 |
| 7 | ANTIRRHINUM MAJUS | 406 | 59% | 4% | 452 |
| 8 | CONTROLLING FLOWER DEVELOPMENT | 325 | 92% | 1% | 128 |
| 9 | APETALA1 | 293 | 83% | 1% | 167 |
| 10 | PISTILLATA | 277 | 92% | 1% | 108 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CURRENT OPINION IN PLANT BIOLOGY | 22 | 11% | 2% | 179 |
| 2 | MOBILE DNA | 9 | 25% | 0% | 30 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| RNA-directed DNA methylation: an epigenetic pathway of increasing complexity | 2014 | 59 | 151 | 86% |
| Transgenerational Epigenetic Inheritance: Myths and Mechanisms | 2014 | 77 | 117 | 50% |
| MicroRNA: a new target for improving plant tolerance to abiotic stress | 2015 | 6 | 102 | 71% |
| The Diversity, Biogenesis, and Activities of Endogenous Silencing Small RNAs in Arabidopsis | 2014 | 47 | 144 | 75% |
| Origin, Biogenesis, and Activity of Plant MicroRNAs | 2009 | 717 | 110 | 67% |
| MicroRNA-Based Biotechnology for Plant Improvement | 2015 | 5 | 233 | 72% |
| Classification and Comparison of Small RNAs from Plants | 2013 | 105 | 121 | 81% |
| The genetic basis of flowering responses to seasonal cues | 2012 | 161 | 129 | 78% |
| Establishing, maintaining and modifying DNA methylation patterns in plants and animals | 2010 | 690 | 179 | 41% |
| Functions of microRNAs in plant stress responses | 2012 | 141 | 76 | 74% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOL PLANT GENET | 33 | 63% | 0.3% | 33 |
| 2 | INTEGRAT GENOME BIOL | 28 | 26% | 0.8% | 92 |
| 3 | MTT BI PLANT GENOM | 27 | 70% | 0.2% | 23 |
| 4 | ZURICH BASEL PLANT SCI | 24 | 27% | 0.7% | 79 |
| 5 | GREGOR MENDEL MOL PLANT BIOL | 23 | 40% | 0.4% | 45 |
| 6 | ABT MOL PFLANZENGENET | 21 | 75% | 0.1% | 15 |
| 7 | MOL EPIGENET MOE | 15 | 42% | 0.2% | 27 |
| 8 | CHROMATIN REPROD GRP | 15 | 88% | 0.1% | 7 |
| 9 | LIFE SCI CORE CURRICULUM | 15 | 88% | 0.1% | 7 |
| 10 | SHANGHAI PLANT ST S BIOL | 14 | 36% | 0.3% | 31 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000027027 | GRAVITROPISM//AUXIN//AUXIN TRANSPORT |
| 2 | 0.0000021848 | DIOECIOUS PLANT//PLANT DEV GENET//SILENE LATIFOLIA |
| 3 | 0.0000015962 | GIBBERELLIN//GIBBERELLIN BIOSYNTHESIS//GIBBERELLINS |
| 4 | 0.0000015833 | DEHYDRIN//ABIOTIC STRESS//DREB |
| 5 | 0.0000012612 | RICE//JAPANESE JOURNAL OF CROP SCIENCE//ORYZA SATIVA |
| 6 | 0.0000011433 | CROWN GALL//PHTHORIMAEA OPERCULELLA//AGROBACTERIUM VITIS |
| 7 | 0.0000010859 | HUMAN ENDOGENOUS RETROVIRUS//ADAR//ENDOGENOUS RETROVIRUS |
| 8 | 0.0000009847 | PLASMODIOPHORA BRASSICAE//CLUBROOT//R ESEED IMPROVEMENT |
| 9 | 0.0000009615 | CHROMOSOMA//SYNAPTONEMAL COMPLEX//MEIOSIS |
| 10 | 0.0000009473 | NEW ZEALAND FLORA//NEW ZEALAND JOURNAL OF BOTANY//GENOME SIZE |