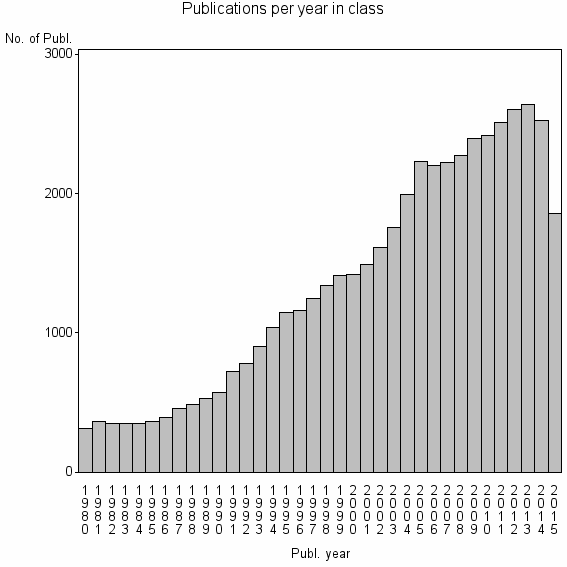
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 204 | 48455 | 42.8 | 75% |
Classes in level above (level 4) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 0 | 3633673 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//MED |
Classes in level below (level 2) |
| ID, lev. below |
Publications | Label for level below |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
| 1035 | 9679 | MOLTEN GLOBULE//APOMYOGLOBIN//PROTEIN FORMULATION |
| 2281 | 4192 | ANGIOGENIN//RIBONUCLEASE//RIBONUCLEASE INHIBITOR |
| 2974 | 2151 | ARGININE METHYLATION//CARM1//ISOASPARTATE |
| 3374 | 1208 | GOOSE TYPE LYSOZYME//G TYPE LYSOZYME//I TYPE LYSOZYME |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN FOLDING | Author keyword | 1132 | 39% | 5% | 2256 |
| 2 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | Journal | 1082 | 38% | 5% | 2289 |
| 3 | PROTEIN STRUCTURE PREDICTION | Author keyword | 707 | 69% | 1% | 609 |
| 4 | PSEUDO AMINO ACID COMPOSITION | Author keyword | 329 | 94% | 0% | 119 |
| 5 | PROTEIN SCIENCE | Journal | 300 | 21% | 3% | 1261 |
| 6 | PROTEIN STABILITY | Author keyword | 231 | 29% | 1% | 670 |
| 7 | BAKER CHEM CHEM BIOL | Address | 223 | 68% | 0% | 193 |
| 8 | MOLTEN GLOBULE | Author keyword | 218 | 53% | 1% | 286 |
| 9 | ANGIOGENIN | Author keyword | 197 | 65% | 0% | 187 |
| 10 | FOLD RECOGNITION | Author keyword | 195 | 69% | 0% | 164 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN FOLDING | 1132 | 39% | 5% | 2256 | Search PROTEIN+FOLDING | Search PROTEIN+FOLDING |
| 2 | PROTEIN STRUCTURE PREDICTION | 707 | 69% | 1% | 609 | Search PROTEIN+STRUCTURE+PREDICTION | Search PROTEIN+STRUCTURE+PREDICTION |
| 3 | PSEUDO AMINO ACID COMPOSITION | 329 | 94% | 0% | 119 | Search PSEUDO+AMINO+ACID+COMPOSITION | Search PSEUDO+AMINO+ACID+COMPOSITION |
| 4 | PROTEIN STABILITY | 231 | 29% | 1% | 670 | Search PROTEIN+STABILITY | Search PROTEIN+STABILITY |
| 5 | MOLTEN GLOBULE | 218 | 53% | 1% | 286 | Search MOLTEN+GLOBULE | Search MOLTEN+GLOBULE |
| 6 | ANGIOGENIN | 197 | 65% | 0% | 187 | Search ANGIOGENIN | Search ANGIOGENIN |
| 7 | FOLD RECOGNITION | 195 | 69% | 0% | 164 | Search FOLD+RECOGNITION | Search FOLD+RECOGNITION |
| 8 | RIBONUCLEASE A | 172 | 66% | 0% | 160 | Search RIBONUCLEASE+A | Search RIBONUCLEASE+A |
| 9 | RIBONUCLEASE | 163 | 44% | 1% | 285 | Search RIBONUCLEASE | Search RIBONUCLEASE |
| 10 | PROTEIN STRUCTURE | 155 | 20% | 1% | 713 | Search PROTEIN+STRUCTURE | Search PROTEIN+STRUCTURE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GLOBULAR PROTEINS | 1355 | 48% | 4% | 2099 |
| 2 | CHYMOTRYPSIN INHIBITOR 2 | 631 | 81% | 1% | 384 |
| 3 | FOLD RECOGNITION | 626 | 70% | 1% | 515 |
| 4 | CONTACT ORDER | 527 | 83% | 1% | 296 |
| 5 | RIBONUCLEASE A | 502 | 49% | 2% | 740 |
| 6 | FUNCTIONAL DOMAIN COMPOSITION | 498 | 89% | 0% | 221 |
| 7 | STRUCTURE PREDICTION | 435 | 33% | 2% | 1085 |
| 8 | MOLTEN GLOBULE STATE | 431 | 51% | 1% | 601 |
| 9 | AMINO ACID COMPOSITION | 424 | 40% | 2% | 838 |
| 10 | ENZYME SUBFAMILY CLASSES | 422 | 97% | 0% | 115 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 1082 | 38% | 5% | 2289 |
| 2 | PROTEIN SCIENCE | 300 | 21% | 3% | 1261 |
| 3 | PROTEIN ENGINEERING | 133 | 23% | 1% | 510 |
| 4 | BIOINFORMATICS | 126 | 11% | 2% | 1070 |
| 5 | JOURNAL OF COMPUTATIONAL CHEMISTRY | 122 | 14% | 2% | 806 |
| 6 | JOURNAL OF CHEMICAL THEORY AND COMPUTATION | 109 | 16% | 1% | 622 |
| 7 | FOLDING & DESIGN | 107 | 62% | 0% | 110 |
| 8 | BMC BIOINFORMATICS | 77 | 10% | 1% | 691 |
| 9 | CURRENT OPINION IN STRUCTURAL BIOLOGY | 63 | 17% | 1% | 347 |
| 10 | PROTEINS-STRUCTURE FUNCTION AND GENETICS | 59 | 31% | 0% | 160 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| CHARMM: The Biomolecular Simulation Program | 2009 | 2048 | 616 | 45% |
| Intrinsically disordered proteins in cellular signalling and regulation | 2015 | 17 | 138 | 54% |
| The Amber biomolecular simulation programs | 2005 | 2779 | 124 | 52% |
| Some remarks on protein attribute prediction and pseudo amino acid composition | 2011 | 329 | 197 | 90% |
| iRSpot-TNCPseAAC: Identify Recombination Spots with Trinucleotide Composition and Pseudo Amino Acid Components | 2014 | 48 | 110 | 88% |
| All-atom empirical potential for molecular modeling and dynamics studies of proteins | 1998 | 6458 | 78 | 33% |
| Locating proteins in the cell using TargetP, SignalP and related tools | 2007 | 1418 | 110 | 41% |
| Interfaces and the driving force of hydrophobic assembly | 2005 | 1314 | 46 | 72% |
| The ensemble nature of allostery | 2014 | 69 | 91 | 49% |
| Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences | 2008 | 586 | 93 | 68% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BAKER CHEM CHEM BIOL | 223 | 68% | 0.4% | 193 |
| 2 | ENGN PROT | 149 | 98% | 0.1% | 40 |
| 3 | BIOL RUMENTAT | 124 | 49% | 0.4% | 186 |
| 4 | COMPUTAT BIOL BIOINFORMAT | 111 | 27% | 0.7% | 352 |
| 5 | SACKLER MOL MED | 102 | 48% | 0.3% | 155 |
| 6 | COMPUTAT MUTAT PROJECT | 82 | 92% | 0.1% | 33 |
| 7 | CHIM BIOPHYS | 65 | 47% | 0.2% | 102 |
| 8 | CUBIC | 60 | 72% | 0.1% | 48 |
| 9 | STUDY SYST BIOL | 52 | 65% | 0.1% | 50 |
| 10 | BIOMOL STRUCT MODELLING UNIT | 51 | 70% | 0.1% | 42 |
Related classes at same level (level 3) |