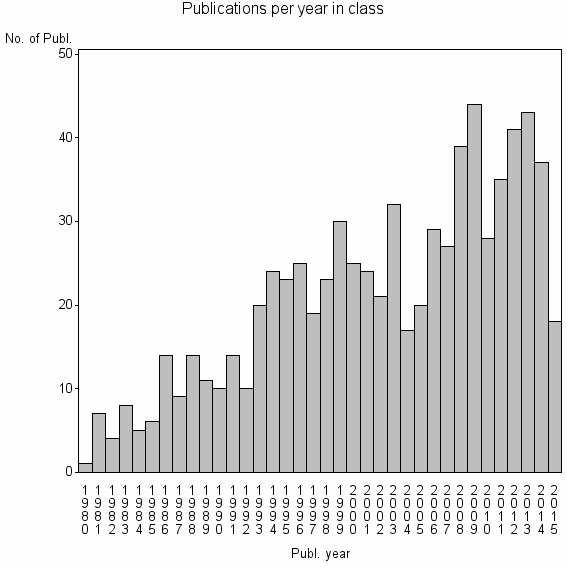
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 13683 | 757 | 36.7 | 72% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2326 | 4027 | ALANINE RACEMASE//TYROSINE PHENOL LYASE//MURD |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | ALANINE RACEMASE | Author keyword | 38 | 56% | 6% | 46 |
| 2 | MURD | Author keyword | 31 | 92% | 2% | 12 |
| 3 | GLUTAMATE RACEMASE | Author keyword | 25 | 70% | 3% | 21 |
| 4 | L AMINO ACID LIGASE | Author keyword | 21 | 90% | 1% | 9 |
| 5 | MUR LIGASES | Author keyword | 20 | 100% | 1% | 9 |
| 6 | MURC | Author keyword | 15 | 68% | 2% | 13 |
| 7 | MURB | Author keyword | 12 | 86% | 1% | 6 |
| 8 | MURD LIGASE | Author keyword | 12 | 86% | 1% | 6 |
| 9 | MUR LIGASE | Author keyword | 11 | 100% | 1% | 6 |
| 10 | PEPTIDOGLYCAN BIOSYNTHESIS | Author keyword | 9 | 44% | 2% | 15 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | ALANINE RACEMASE | 38 | 56% | 6% | 46 | Search ALANINE+RACEMASE | Search ALANINE+RACEMASE |
| 2 | MURD | 31 | 92% | 2% | 12 | Search MURD | Search MURD |
| 3 | GLUTAMATE RACEMASE | 25 | 70% | 3% | 21 | Search GLUTAMATE+RACEMASE | Search GLUTAMATE+RACEMASE |
| 4 | L AMINO ACID LIGASE | 21 | 90% | 1% | 9 | Search L+AMINO+ACID+LIGASE | Search L+AMINO+ACID+LIGASE |
| 5 | MUR LIGASES | 20 | 100% | 1% | 9 | Search MUR+LIGASES | Search MUR+LIGASES |
| 6 | MURC | 15 | 68% | 2% | 13 | Search MURC | Search MURC |
| 7 | MURB | 12 | 86% | 1% | 6 | Search MURB | Search MURB |
| 8 | MURD LIGASE | 12 | 86% | 1% | 6 | Search MURD+LIGASE | Search MURD+LIGASE |
| 9 | MUR LIGASE | 11 | 100% | 1% | 6 | Search MUR+LIGASE | Search MUR+LIGASE |
| 10 | PEPTIDOGLYCAN BIOSYNTHESIS | 9 | 44% | 2% | 15 | Search PEPTIDOGLYCAN+BIOSYNTHESIS | Search PEPTIDOGLYCAN+BIOSYNTHESIS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ADDING ENZYME | 67 | 75% | 6% | 48 |
| 2 | ALANYL D GLUTAMATE | 49 | 94% | 2% | 17 |
| 3 | PHOSPHINATE INHIBITORS | 49 | 94% | 2% | 17 |
| 4 | L ALANINE LIGASE | 41 | 85% | 3% | 22 |
| 5 | ANTIBIOTIC FOSFOMYCIN | 35 | 64% | 5% | 35 |
| 6 | ACID ADDING ENZYME | 31 | 74% | 3% | 23 |
| 7 | PEPTIDOGLYCAN BIOSYNTHESIS | 29 | 29% | 11% | 85 |
| 8 | ALANINE ADDING ENZYME | 26 | 87% | 2% | 13 |
| 9 | DDLA GENE | 23 | 79% | 2% | 15 |
| 10 | N ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE | 23 | 86% | 2% | 12 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Cytoplasmic steps of peptidoglycan biosynthesis | 2008 | 139 | 350 | 66% |
| Structure and function of the Mur enzymes: development of novel inhibitors | 2003 | 128 | 57 | 88% |
| Recent advances in the formation of the bacterial peptidoglycan monomer unit | 2001 | 224 | 254 | 53% |
| Bacterial cell wall assembly: still an attractive antibacterial target | 2011 | 46 | 73 | 41% |
| Structure, function and dynamics in the mur family of bacterial cell wall ligases | 2006 | 57 | 58 | 60% |
| The biology of Mur ligases as an antibacterial target | 2014 | 2 | 81 | 69% |
| Prospects for new antibiotics: a molecule-centered perspective | 2014 | 24 | 202 | 10% |
| Challenges of Antibacterial Discovery | 2011 | 199 | 334 | 10% |
| Development of novel inhibitors targeting intracellular steps of peptidoglycan biosynthesis | 2007 | 38 | 149 | 56% |
| Structure-based design approaches to cell wall biosynthesis inhibitors | 2003 | 47 | 60 | 72% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | IBBMC | 7 | 19% | 4.2% | 32 |
| 2 | ENVELOPPES BACTERIENNES ANTIBIOT | 5 | 26% | 2.0% | 15 |
| 3 | VMKV ENGN | 1 | 31% | 0.5% | 4 |
| 4 | INFECT DISCOVERY | 1 | 18% | 0.8% | 6 |
| 5 | CNRS URA 1131 | 1 | 40% | 0.3% | 2 |
| 6 | RECH FONCT STRUCT INGN PROT | 1 | 13% | 0.9% | 7 |
| 7 | FAK FARM | 1 | 19% | 0.5% | 4 |
| 8 | CHEM SCI INFECT DIS | 1 | 33% | 0.3% | 2 |
| 9 | CRISTALLOG MACROMOL | 1 | 12% | 0.8% | 6 |
| 10 | AGRON HORTICULTURAL SCI | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000124323 | ASPARTATE AMINOTRANSFERASE//AROMATIC AMINO ACID AMINOTRANSFERASE//PYRIDOXAL 5 PHOSPHATE |
| 2 | 0.0000113295 | MANDELATE RACEMASE//ENOLASE SUPERFAMILY//MANDELATE RACEMASE EC 5122 |
| 3 | 0.0000109827 | TRANSLOCASE I//MRAY//CAPRAZAMYCIN |
| 4 | 0.0000101000 | UMR8041//USM0502//MUREIN |
| 5 | 0.0000096040 | ESSENTIAL GENES//SUBTRACTIVE GENOMICS//ESSENTIAL GENE |
| 6 | 0.0000088523 | D AMINO ACID OXIDASE//D SERINE//SERINE RACEMASE |
| 7 | 0.0000082106 | FOSFOMYCIN RESISTANCE//FOSFOMYCIN//AREA PROD PORCINA |
| 8 | 0.0000076545 | DIHYDRODIPICOLINATE SYNTHASE//ASPARTATE KINASE//THREONINE SYNTHASE |
| 9 | 0.0000076387 | GLUCOSAMINE 6 PHOSPHATE SYNTHASE//GLUCOSAMINE 6P SYNTHASE//GLUCOSAMINE 6 PHOSPHATE DEAMINASE |
| 10 | 0.0000075392 | DIBROMANTIN//1 3 DIBROMO 5 5 DIMETHYL 2 4 IMIDAZOLIDINEDIONE 77 48 5//BETA KETO CARBONYL COMPOUNDS |