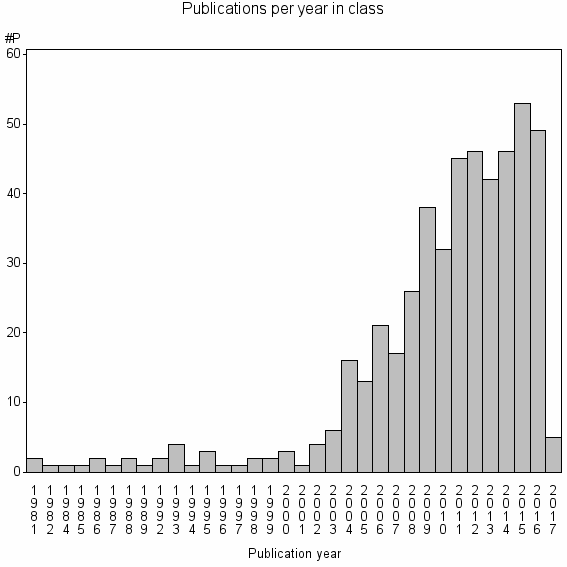
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 19752 | 490 | 40.3 | 78% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 3 | 4 | PLANT SCIENCES//AGRONOMY//FOOD SCIENCE & TECHNOLOGY | 1882812 |
| 300 | 3 | PHOTOSYSTEM II//PHOTOSYNTHESIS RESEARCH//REACTION CENTER | 41101 |
| 2089 | 2 | MACROMOLECULAR CROWDING//LEVY WALK//CORRELATED RANDOM WALK | 5197 |
| 19752 | 1 | NARROW ESCAPE//NONSPECIFIC PROTEIN DNA INTERACTIONS//TARGET SEARCH PROCESS | 490 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | NARROW ESCAPE | authKW | 538584 | 2% | 79% | 11 |
| 2 | NONSPECIFIC PROTEIN DNA INTERACTIONS | authKW | 186948 | 1% | 100% | 3 |
| 3 | TARGET SEARCH PROCESS | authKW | 186948 | 1% | 100% | 3 |
| 4 | MEAN FIRST PASSAGE TIME | authKW | 177102 | 4% | 15% | 19 |
| 5 | NARROW ESCAPE PROBLEM | authKW | 140210 | 1% | 75% | 3 |
| 6 | NEUMANN GREENS FUNCTION | authKW | 140210 | 1% | 75% | 3 |
| 7 | ASYMPTOTIC ANALYSIS OF PDES | authKW | 124632 | 0% | 100% | 2 |
| 8 | DNA SEARCH | authKW | 124632 | 0% | 100% | 2 |
| 9 | GAUSS CONSTANT | authKW | 124632 | 0% | 100% | 2 |
| 10 | INTERMITTENT SEARCH STRATEGIES | authKW | 124632 | 0% | 100% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Physics, Mathematical | 3491 | 24% | 0% | 117 |
| 2 | Physics, Fluids & Plasmas | 1148 | 12% | 0% | 60 |
| 3 | Biophysics | 701 | 14% | 0% | 71 |
| 4 | Biochemistry & Molecular Biology | 569 | 31% | 0% | 151 |
| 5 | Physics, Multidisciplinary | 249 | 12% | 0% | 58 |
| 6 | Mathematical & Computational Biology | 232 | 4% | 0% | 21 |
| 7 | Physics, Atomic, Molecular & Chemical | 202 | 9% | 0% | 45 |
| 8 | Chemistry, Physical | 128 | 14% | 0% | 67 |
| 9 | Mathematics, Applied | 66 | 6% | 0% | 29 |
| 10 | Mathematics, Interdisciplinary Applications | 45 | 3% | 0% | 13 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MA1 | 124632 | 0% | 100% | 2 |
| 2 | MUSEUM HIST NATU565 | 124632 | 0% | 100% | 2 |
| 3 | GRP PL MATH COMPUTAT BIOL | 99701 | 1% | 40% | 4 |
| 4 | BIOL NETWORKS GRP | 62316 | 0% | 100% | 1 |
| 5 | CASE COURIER 121 | 62316 | 0% | 100% | 1 |
| 6 | FUNCT IMAGING TRANSCRIPTIBENS | 62316 | 0% | 100% | 1 |
| 7 | GRENOBLE LITEN DTNM L2T | 62316 | 0% | 100% | 1 |
| 8 | JEAN PENIN | 62316 | 0% | 100% | 1 |
| 9 | JEAN PERRIN FRE 3231 | 62316 | 0% | 100% | 1 |
| 10 | JUNIOR FELLOW PROGRAM | 62316 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MULTISCALE MODELING & SIMULATION | 11321 | 2% | 2% | 12 |
| 2 | PHYSICAL REVIEW E | 4185 | 12% | 0% | 60 |
| 3 | BIOPHYSICAL JOURNAL | 3886 | 7% | 0% | 35 |
| 4 | JOURNAL OF PHYSICS A-MATHEMATICAL AND THEORETICAL | 2890 | 4% | 0% | 20 |
| 5 | PHYSICAL BIOLOGY | 2694 | 1% | 1% | 6 |
| 6 | JOURNAL OF STATISTICAL PHYSICS | 2006 | 3% | 0% | 16 |
| 7 | EUROPEAN JOURNAL OF APPLIED MATHEMATICS | 1526 | 1% | 1% | 4 |
| 8 | JOURNAL OF BIOLOGICAL PHYSICS | 1336 | 1% | 1% | 4 |
| 9 | NUCLEIC ACIDS RESEARCH | 1201 | 6% | 0% | 28 |
| 10 | SIAM JOURNAL ON APPLIED MATHEMATICS | 1143 | 2% | 0% | 8 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NARROW ESCAPE | 538584 | 2% | 79% | 11 | Search NARROW+ESCAPE | Search NARROW+ESCAPE |
| 2 | NONSPECIFIC PROTEIN DNA INTERACTIONS | 186948 | 1% | 100% | 3 | Search NONSPECIFIC+PROTEIN+DNA+INTERACTIONS | Search NONSPECIFIC+PROTEIN+DNA+INTERACTIONS |
| 3 | TARGET SEARCH PROCESS | 186948 | 1% | 100% | 3 | Search TARGET+SEARCH+PROCESS | Search TARGET+SEARCH+PROCESS |
| 4 | MEAN FIRST PASSAGE TIME | 177102 | 4% | 15% | 19 | Search MEAN+FIRST+PASSAGE+TIME | Search MEAN+FIRST+PASSAGE+TIME |
| 5 | NARROW ESCAPE PROBLEM | 140210 | 1% | 75% | 3 | Search NARROW+ESCAPE+PROBLEM | Search NARROW+ESCAPE+PROBLEM |
| 6 | NEUMANN GREENS FUNCTION | 140210 | 1% | 75% | 3 | Search NEUMANN+GREENS+FUNCTION | Search NEUMANN+GREENS+FUNCTION |
| 7 | ASYMPTOTIC ANALYSIS OF PDES | 124632 | 0% | 100% | 2 | Search ASYMPTOTIC+ANALYSIS+OF+PDES | Search ASYMPTOTIC+ANALYSIS+OF+PDES |
| 8 | DNA SEARCH | 124632 | 0% | 100% | 2 | Search DNA+SEARCH | Search DNA+SEARCH |
| 9 | GAUSS CONSTANT | 124632 | 0% | 100% | 2 | Search GAUSS+CONSTANT | Search GAUSS+CONSTANT |
| 10 | INTERMITTENT SEARCH STRATEGIES | 124632 | 0% | 100% | 2 | Search INTERMITTENT+SEARCH+STRATEGIES | Search INTERMITTENT+SEARCH+STRATEGIES |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | TABAKA, M , BURDZY, K , HOLYST, R , (2015) METHOD FOR THE ANALYSIS OF CONTRIBUTION OF SLIDING AND HOPPING TO A FACILITATED DIFFUSION OF DNA-BINDING PROTEIN: APPLICATION TO IN VIVO DATA.PHYSICAL REVIEW E. VOL. 92. ISSUE 2. P. - | 59 | 92% | 1 |
| 2 | VEKSLER, A , KOLOMEISKY, AB , (2013) SPEED-SELECTIVITY PARADOX IN THE PROTEIN SEARCH FOR TARGETS ON DNA: IS IT REAL OR NOT?.JOURNAL OF PHYSICAL CHEMISTRY B. VOL. 117. ISSUE 42. P. 12695-12701 | 48 | 87% | 17 |
| 3 | NIRANJANI, G , MURUGAN, R , (2016) GENERALIZED THEORY ON THE MECHANISM OF SITE-SPECIFIC DNA-PROTEIN INTERACTIONS.JOURNAL OF STATISTICAL MECHANICS-THEORY AND EXPERIMENT. VOL. . ISSUE . P. - | 49 | 83% | 0 |
| 4 | KOLOMEISKY, AB , (2011) PHYSICS OF PROTEIN-DNA INTERACTIONS: MECHANISMS OF FACILITATED TARGET SEARCH.PHYSICAL CHEMISTRY CHEMICAL PHYSICS. VOL. 13. ISSUE 6. P. 2088 -2095 | 40 | 85% | 64 |
| 5 | MAHMUTOVIC, A , BERG, OG , ELF, J , (2015) WHAT MATTERS FOR LAC REPRESSOR SEARCH IN VIVO-SLIDING, HOPPING, INTERSEGMENT TRANSFER, CROWDING ON DNA OR RECOGNITION?.NUCLEIC ACIDS RESEARCH. VOL. 43. ISSUE 7. P. 3454 -3464 | 39 | 80% | 10 |
| 6 | MARCOVITZ, A , LEVY, Y , (2013) OBSTACLES MAY FACILITATE AND DIRECT DNA SEARCH BY PROTEINS.BIOPHYSICAL JOURNAL. VOL. 104. ISSUE 9. P. 2042-2050 | 40 | 83% | 24 |
| 7 | BAUER, M , RASMUSSEN, ES , LOMHOLT, MA , METZLER, R , (2015) REAL SEQUENCE EFFECTS ON THE SEARCH DYNAMICS OF TRANSCRIPTION FACTORS ON DNA.SCIENTIFIC REPORTS. VOL. 5. ISSUE . P. - | 39 | 75% | 12 |
| 8 | SHEINMAN, M , BENICHOU, O , KAFRI, Y , VOITURIEZ, R , (2012) CLASSES OF FAST AND SPECIFIC SEARCH MECHANISMS FOR PROTEINS ON DNA.REPORTS ON PROGRESS IN PHYSICS. VOL. 75. ISSUE 2. P. - | 58 | 53% | 46 |
| 9 | LIU, L , LUO, KF , (2015) DNA-BINDING PROTEIN SEARCHES FOR ITS TARGET: NON-MONOTONIC DEPENDENCE OF THE SEARCH TIME ON THE DENSITY OF ROADBLOCKS BOUND ON THE DNA CHAIN.JOURNAL OF CHEMICAL PHYSICS. VOL. 142. ISSUE 12. P. - | 35 | 95% | 2 |
| 10 | KREPEL, D , GOMEZ, D , KLUMPP, S , LEVY, Y , (2016) MECHANISM OF FACILITATED DIFFUSION DURING A DNA SEARCH IN CROWDED ENVIRONMENTS.JOURNAL OF PHYSICAL CHEMISTRY B. VOL. 120. ISSUE 43. P. 11113 -11122 | 46 | 70% | 0 |
Classes with closest relation at Level 1 |