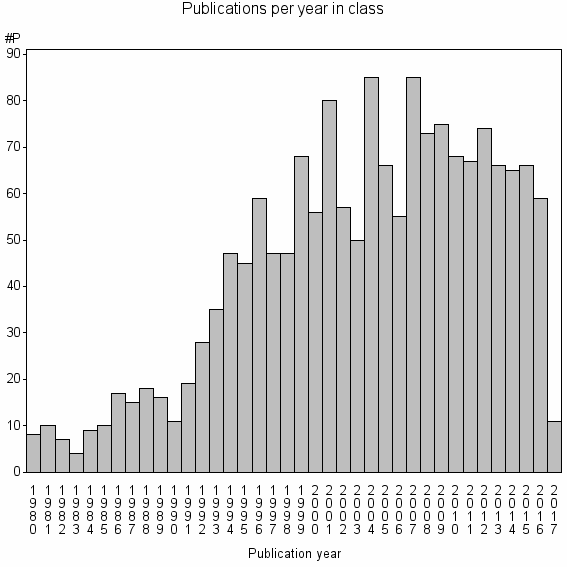
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 5071 | 1678 | 51.2 | 85% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 27 | 3 | DEVELOPMENTAL BIOLOGY//DEVELOPMENT//AGR2 | 109777 |
| 253 | 2 | DROSOPHILA//DEVELOPMENTAL BIOLOGY//DEVELOPMENT | 19712 |
| 5071 | 1 | RNA LOCALIZATION//OSKAR//GERM PLASM | 1678 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RNA LOCALIZATION | authKW | 1028024 | 7% | 50% | 114 |
| 2 | OSKAR | authKW | 812300 | 3% | 89% | 50 |
| 3 | GERM PLASM | authKW | 595879 | 4% | 50% | 66 |
| 4 | PUMILIO | authKW | 591065 | 3% | 69% | 47 |
| 5 | OOGENESIS | authKW | 494983 | 12% | 13% | 207 |
| 6 | MRNA LOCALIZATION | authKW | 385195 | 4% | 32% | 67 |
| 7 | GURKEN | authKW | 345019 | 2% | 59% | 32 |
| 8 | NANOS | authKW | 334747 | 3% | 40% | 46 |
| 9 | DROSOPHILA OOGENESIS | authKW | 268431 | 2% | 49% | 30 |
| 10 | FUSOME | authKW | 250963 | 1% | 69% | 20 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Developmental Biology | 79069 | 44% | 1% | 731 |
| 2 | Cell Biology | 12748 | 45% | 0% | 748 |
| 3 | Biochemistry & Molecular Biology | 2423 | 34% | 0% | 566 |
| 4 | Genetics & Heredity | 1167 | 12% | 0% | 204 |
| 5 | Anatomy & Morphology | 285 | 2% | 0% | 33 |
| 6 | Biology | 138 | 3% | 0% | 55 |
| 7 | Reproductive Biology | 82 | 2% | 0% | 29 |
| 8 | Evolutionary Biology | 27 | 1% | 0% | 21 |
| 9 | Biochemical Research Methods | 26 | 2% | 0% | 39 |
| 10 | Biophysics | 22 | 3% | 0% | 42 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOMED COMPUTAT VISUALIZAT | 109175 | 0% | 100% | 6 |
| 2 | GERMLINE DEV | 79101 | 1% | 43% | 10 |
| 3 | SKIRBALL HOWARD HUGHES MED | 54587 | 0% | 100% | 3 |
| 4 | GENE EXPT | 30991 | 1% | 15% | 11 |
| 5 | GRP BIOMOL NMR | 24260 | 0% | 67% | 2 |
| 6 | PROGRAMA GULBENKIAN DOUTORAMENTO BIOL MED | 24260 | 0% | 67% | 2 |
| 7 | STELLAR CHANCE S 709B | 24260 | 0% | 67% | 2 |
| 8 | STEM CELL CONSORTIUM | 24260 | 0% | 67% | 2 |
| 9 | DEV BIOL INITIAT | 22737 | 0% | 25% | 5 |
| 10 | SECT MOL CELL DEV BIOL | 19869 | 1% | 6% | 19 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DEVELOPMENT | 56370 | 12% | 2% | 198 |
| 2 | MECHANISMS OF DEVELOPMENT | 19575 | 3% | 2% | 58 |
| 3 | DEVELOPMENTAL BIOLOGY | 17447 | 7% | 1% | 115 |
| 4 | DEVELOPMENT GROWTH & DIFFERENTIATION | 14430 | 3% | 2% | 45 |
| 5 | CURRENT BIOLOGY | 9409 | 4% | 1% | 68 |
| 6 | RNA | 9246 | 2% | 1% | 36 |
| 7 | DEVELOPMENTAL CELL | 5906 | 2% | 1% | 29 |
| 8 | GENES & DEVELOPMENT | 4888 | 3% | 1% | 45 |
| 9 | DEVELOPMENT GENES AND EVOLUTION | 4147 | 1% | 1% | 17 |
| 10 | CURRENT TOPICS IN DEVELOPMENTAL BIOLOGY | 3469 | 1% | 1% | 14 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA LOCALIZATION | 1028024 | 7% | 50% | 114 | Search RNA+LOCALIZATION | Search RNA+LOCALIZATION |
| 2 | OSKAR | 812300 | 3% | 89% | 50 | Search OSKAR | Search OSKAR |
| 3 | GERM PLASM | 595879 | 4% | 50% | 66 | Search GERM+PLASM | Search GERM+PLASM |
| 4 | PUMILIO | 591065 | 3% | 69% | 47 | Search PUMILIO | Search PUMILIO |
| 5 | OOGENESIS | 494983 | 12% | 13% | 207 | Search OOGENESIS | Search OOGENESIS |
| 6 | MRNA LOCALIZATION | 385195 | 4% | 32% | 67 | Search MRNA+LOCALIZATION | Search MRNA+LOCALIZATION |
| 7 | GURKEN | 345019 | 2% | 59% | 32 | Search GURKEN | Search GURKEN |
| 8 | NANOS | 334747 | 3% | 40% | 46 | Search NANOS | Search NANOS |
| 9 | DROSOPHILA OOGENESIS | 268431 | 2% | 49% | 30 | Search DROSOPHILA+OOGENESIS | Search DROSOPHILA+OOGENESIS |
| 10 | FUSOME | 250963 | 1% | 69% | 20 | Search FUSOME | Search FUSOME |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LEHMANN, R , (2016) GERM PLASM BIOGENESIS-AN OSKAR-CENTRIC PERSPECTIVE.ESSAYS ON DEVELOPMENTAL BIOLOGY, PT A. VOL. 116. ISSUE . P. 679 -+ | 107 | 80% | 3 |
| 2 | ST JOHNSTON, D , (2005) MOVING MESSAGES: THE INTRACELLULAR LOCALIZATION OF MRNAS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 6. ISSUE 5. P. 363-375 | 127 | 69% | 305 |
| 3 | KUGLER, JM , LASKO, P , (2009) LOCALIZATION, ANCHORING AND TRANSLATIONAL CONTROL OF OSKAR, GURKEN, BICOID AND NANOS MRNA DURING DROSOPHILA OOGENESIS.FLY. VOL. 3. ISSUE 1. P. 15 -28 | 100 | 81% | 57 |
| 4 | KLOC, M , ETKIN, LD , (2005) RNA LOCALIZATION MECHANISMS IN OOCYTES.JOURNAL OF CELL SCIENCE. VOL. 118. ISSUE 2. P. 269-282 | 119 | 71% | 83 |
| 5 | JOHNSTONE, O , LASKO, P , (2001) TRANSLATIONAL REGULATION AND RNA LOCALIZATION IN DROSOPHILA OOCYTES AND EMBRYOS.ANNUAL REVIEW OF GENETICS. VOL. 35. ISSUE . P. 365-406 | 140 | 56% | 193 |
| 6 | BASHIRULLAH, A , COOPERSTOCK, RL , LIPSHITZ, HD , (1998) RNA LOCALIZATION IN DEVELOPMENT.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 67. ISSUE . P. 335 -394 | 134 | 57% | 249 |
| 7 | SHAHBABIAN, K , CHARTRAND, P , (2012) CONTROL OF CYTOPLASMIC MRNA LOCALIZATION.CELLULAR AND MOLECULAR LIFE SCIENCES. VOL. 69. ISSUE 4. P. 535 -552 | 110 | 59% | 9 |
| 8 | LASKO, P , (2012) MRNA LOCALIZATION AND TRANSLATIONAL CONTROL IN DROSOPHILA OOGENESIS.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 4. ISSUE 10. P. - | 90 | 70% | 15 |
| 9 | JANSEN, RP , NIESSING, D , (2012) ASSEMBLY OF MRNA-PROTEIN COMPLEXES FOR DIRECTIONAL MRNA TRANSPORT IN EUKARYOTES - AN OVERVIEW.CURRENT PROTEIN & PEPTIDE SCIENCE. VOL. 13. ISSUE 4. P. 284 -293 | 82 | 72% | 15 |
| 10 | PALACIOS, IM , JOHNSTON, DS , (2001) GETTING THE MESSAGE ACROSS: THE INTRACELLULAR LOCALIZATION OF MRNAS IN HIGHER EUKARYOTES.ANNUAL REVIEW OF CELL AND DEVELOPMENTAL BIOLOGY. VOL. 17. ISSUE . P. 569 -614 | 138 | 51% | 153 |
Classes with closest relation at Level 1 |