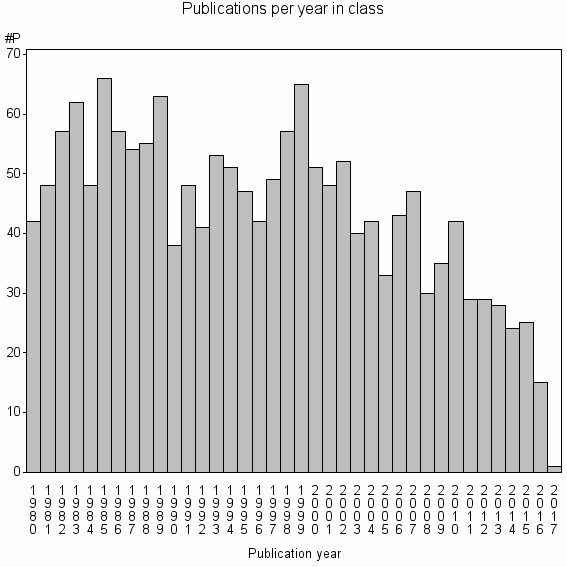
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 5203 | 1657 | 40.0 | 69% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 3 | 4 | PLANT SCIENCES//AGRONOMY//FOOD SCIENCE & TECHNOLOGY | 1882812 |
| 7 | 3 | PLANT SCIENCES//PLANT PHYSIOLOGY//ARABIDOPSIS | 149859 |
| 1802 | 2 | GLUTAMINE SYNTHETASE//NITRATE REDUCTASE//GLUTAMATE SYNTHASE | 6286 |
| 5203 | 1 | SIGMA54//PII PROTEIN//GLNB | 1657 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | SIGMA54 | authKW | 701249 | 4% | 61% | 62 |
| 2 | PII PROTEIN | authKW | 327612 | 2% | 66% | 27 |
| 3 | GLNB | authKW | 318504 | 1% | 79% | 22 |
| 4 | NIFA PROTEIN | authKW | 276396 | 1% | 100% | 15 |
| 5 | GLNK | authKW | 272973 | 1% | 74% | 20 |
| 6 | NIFA | authKW | 258246 | 2% | 48% | 29 |
| 7 | HERBASPIRILLUM SEROPEDICAE | authKW | 186533 | 2% | 38% | 27 |
| 8 | NTRC | authKW | 179763 | 1% | 51% | 19 |
| 9 | NITROGEN REGULATION | authKW | 179725 | 2% | 26% | 38 |
| 10 | STUDY NITROGEN FIXAT | address | 171766 | 1% | 55% | 17 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Microbiology | 25142 | 51% | 0% | 848 |
| 2 | Biochemistry & Molecular Biology | 5026 | 46% | 0% | 769 |
| 3 | Genetics & Heredity | 899 | 11% | 0% | 181 |
| 4 | Biophysics | 205 | 5% | 0% | 85 |
| 5 | Cell Biology | 149 | 7% | 0% | 115 |
| 6 | Biology | 128 | 3% | 0% | 53 |
| 7 | Biotechnology & Applied Microbiology | 93 | 5% | 0% | 78 |
| 8 | Biochemical Research Methods | 18 | 2% | 0% | 35 |
| 9 | Plant Sciences | 13 | 3% | 0% | 49 |
| 10 | Soil Science | 9 | 1% | 0% | 13 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | STUDY NITROGEN FIXAT | 171766 | 1% | 55% | 17 |
| 2 | NACL CIENCIA TECNOL FIXACAO BIOL NITROGENI | 106603 | 1% | 64% | 9 |
| 3 | INTEGRAT BIOSCI DEGREE PROGRAM CHEM BIOL | 55279 | 0% | 100% | 3 |
| 4 | SCI TECHNOL BIOL NITROGEN FIXAT | 55279 | 0% | 100% | 3 |
| 5 | AD TAT MICROORGANISMS | 49134 | 0% | 67% | 4 |
| 6 | RECONOCIMIENTO MOL BIOESTRUCTURA | 32754 | 0% | 44% | 4 |
| 7 | STRUCT BIOL MOL BIOSCI | 24567 | 0% | 67% | 2 |
| 8 | URA 1300UNITE PHYSIOL CELLULAIRE | 24567 | 0% | 67% | 2 |
| 9 | GRP SYMBIOSIS | 23688 | 0% | 43% | 3 |
| 10 | AGROMICROBIOL OURCE PL AGR MINIST | 18426 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF BACTERIOLOGY | 59217 | 20% | 1% | 326 |
| 2 | MOLECULAR MICROBIOLOGY | 15730 | 6% | 1% | 101 |
| 3 | ARCHIVES OF MICROBIOLOGY | 11291 | 4% | 1% | 58 |
| 4 | MOLECULAR AND GENERAL GENETICS | 8410 | 3% | 1% | 47 |
| 5 | FEMS MICROBIOLOGY LETTERS | 7910 | 5% | 1% | 83 |
| 6 | MOLECULAR & GENERAL GENETICS | 7247 | 2% | 1% | 31 |
| 7 | CURRENT TOPICS IN CELLULAR REGULATION | 6075 | 1% | 4% | 9 |
| 8 | JOURNAL OF GENERAL MICROBIOLOGY | 5005 | 2% | 1% | 38 |
| 9 | JOURNAL OF MOLECULAR BIOLOGY | 3563 | 4% | 0% | 68 |
| 10 | MICROBIOLOGY-SGM | 2068 | 2% | 0% | 30 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SIGMA54 | 701249 | 4% | 61% | 62 | Search SIGMA54 | Search SIGMA54 |
| 2 | PII PROTEIN | 327612 | 2% | 66% | 27 | Search PII+PROTEIN | Search PII+PROTEIN |
| 3 | GLNB | 318504 | 1% | 79% | 22 | Search GLNB | Search GLNB |
| 4 | NIFA PROTEIN | 276396 | 1% | 100% | 15 | Search NIFA+PROTEIN | Search NIFA+PROTEIN |
| 5 | GLNK | 272973 | 1% | 74% | 20 | Search GLNK | Search GLNK |
| 6 | NIFA | 258246 | 2% | 48% | 29 | Search NIFA | Search NIFA |
| 7 | HERBASPIRILLUM SEROPEDICAE | 186533 | 2% | 38% | 27 | Search HERBASPIRILLUM+SEROPEDICAE | Search HERBASPIRILLUM+SEROPEDICAE |
| 8 | NTRC | 179763 | 1% | 51% | 19 | Search NTRC | Search NTRC |
| 9 | NITROGEN REGULATION | 179725 | 2% | 26% | 38 | Search NITROGEN+REGULATION | Search NITROGEN+REGULATION |
| 10 | NIFL | 153550 | 1% | 83% | 10 | Search NIFL | Search NIFL |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | HUERGO, LF , CHANDRA, G , MERRICK, M , (2013) PII SIGNAL TRANSDUCTION PROTEINS: NITROGEN REGULATION AND BEYOND.FEMS MICROBIOLOGY REVIEWS. VOL. 37. ISSUE 2. P. 251 -283 | 219 | 78% | 33 |
| 2 | ARCONDEGUY, T , JACK, R , MERRICK, M , (2001) P-II SIGNAL TRANSDUCTION PROTEINS, PIVOTAL PLAYERS IN MICROBIAL NITROGEN CONTROL.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 65. ISSUE 1. P. 80 -+ | 157 | 70% | 271 |
| 3 | BUSH, M , DIXON, R , (2012) THE ROLE OF BACTERIAL ENHANCER BINDING PROTEINS AS SPECIALIZED ACTIVATORS OF SIGMA(54)-DEPENDENT TRANSCRIPTION.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 76. ISSUE 3. P. 497-529 | 133 | 57% | 62 |
| 4 | MERRICK, MJ , EDWARDS, RA , (1995) NITROGEN CONTROL IN BACTERIA.MICROBIOLOGICAL REVIEWS. VOL. 59. ISSUE 4. P. 604-& | 156 | 64% | 420 |
| 5 | HUERGO, LF , PEDROSA, FO , MULLER-SANTOS, M , CHUBATSU, LS , MONTEIRO, RA , MERRICK, M , SOUZA, EM , (2012) P-II SIGNAL TRANSDUCTION PROTEINS: PIVOTAL PLAYERS IN POST-TRANSLATIONAL CONTROL OF NITROGENASE ACTIVITY.MICROBIOLOGY-SGM. VOL. 158. ISSUE . P. 176 -190 | 92 | 85% | 16 |
| 6 | VAN HEESWIJK, WC , WESTERHOFF, HV , BOOGERD, FC , (2013) NITROGEN ASSIMILATION IN ESCHERICHIA COLI: PUTTING MOLECULAR DATA INTO A SYSTEMS PERSPECTIVE.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 77. ISSUE 4. P. 628-695 | 194 | 37% | 22 |
| 7 | LEIGH, JA , DODSWORTH, JA , (2007) NITROGEN REGULATION IN BACTERIA AND ARCHAEA.ANNUAL REVIEW OF MICROBIOLOGY. VOL. 61. ISSUE . P. 349 -377 | 69 | 69% | 168 |
| 8 | ZOU, XX , ZHU, Y , POHLMANN, EL , LI, JL , ZHANG, YP , ROBERTS, GP , (2008) IDENTIFICATION AND FUNCTIONAL CHARACTERIZATION OF NIFA VARIANTS THAT ARE INDEPENDENT OF GLNB ACTIVATION IN THE PHOTOSYNTHETIC BACTERIUM RHODOSPIRILLUM RUBRUM.MICROBIOLOGY-SGM. VOL. 154. ISSUE . P. 2689-2699 | 65 | 89% | 10 |
| 9 | LUDDECKE, J , FORCHHAMMER, K , (2016) SENSORY PROPERTIES OF THE P-II SIGNALLING PROTEIN FAMILY.FEBS JOURNAL. VOL. 283. ISSUE 3. P. 425 -437 | 59 | 71% | 2 |
| 10 | ZHANG, YP , POHLMANN, EL , CONRAD, MC , ROBERTS, GP , (2006) THE POOR GROWTH OF RHODOSPIRILLUM RUBRUM MUTANTS LACKING P-II PROTEINS IS DUE TO AN EXCESS OF GLUTAMINE SYNTHETASE ACTIVITY.MOLECULAR MICROBIOLOGY. VOL. 61. ISSUE 2. P. 497-510 | 69 | 85% | 15 |
Classes with closest relation at Level 1 |