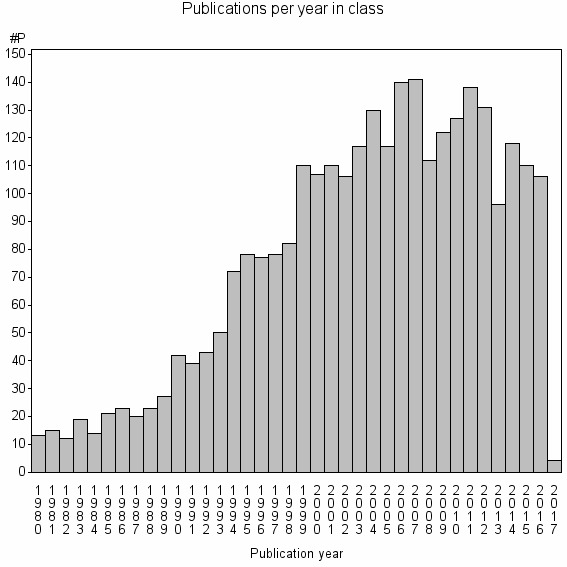
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 1038 | 2890 | 50.0 | 84% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 364 | 3 | TELOMERASE//TELOMERE//C ELEGANS | 34320 |
| 1156 | 2 | C ELEGANS//CAENORHABDITIS ELEGANS//DAF 16 | 9487 |
| 1038 | 1 | C ELEGANS//CAENORHABDITIS ELEGANS//P GRANULES | 2890 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | C ELEGANS | authKW | 1399970 | 19% | 24% | 545 |
| 2 | CAENORHABDITIS ELEGANS | authKW | 460569 | 12% | 13% | 343 |
| 3 | P GRANULES | authKW | 202820 | 1% | 80% | 24 |
| 4 | VULVAL DEVELOPMENT | authKW | 186343 | 1% | 84% | 21 |
| 5 | POP 1 | authKW | 158460 | 1% | 100% | 15 |
| 6 | CELL FATE DECIS | address | 152645 | 1% | 85% | 17 |
| 7 | VULVA DEVELOPMENT | authKW | 147896 | 0% | 100% | 14 |
| 8 | DEVELOPMENTAL BIOLOGY | WoSSC | 137713 | 44% | 1% | 1266 |
| 9 | LIN 12 | authKW | 128033 | 1% | 61% | 20 |
| 10 | HLH 2 | authKW | 119018 | 0% | 87% | 13 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Developmental Biology | 137713 | 44% | 1% | 1266 |
| 2 | Cell Biology | 12427 | 34% | 0% | 986 |
| 3 | Genetics & Heredity | 11759 | 27% | 0% | 784 |
| 4 | Biochemistry & Molecular Biology | 1828 | 24% | 0% | 699 |
| 5 | Biology | 381 | 4% | 0% | 114 |
| 6 | Anatomy & Morphology | 306 | 2% | 0% | 46 |
| 7 | Evolutionary Biology | 76 | 1% | 0% | 43 |
| 8 | Biochemical Research Methods | 63 | 3% | 0% | 74 |
| 9 | Biotechnology & Applied Microbiology | 41 | 3% | 0% | 92 |
| 10 | Mathematical & Computational Biology | 36 | 1% | 0% | 28 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CELL FATE DECIS | 152645 | 1% | 85% | 17 |
| 2 | ABT EVOLUT BIOL | 98249 | 1% | 47% | 20 |
| 3 | MULTICELLULAR ORG | 67607 | 0% | 80% | 8 |
| 4 | 3944 | 37552 | 0% | 44% | 8 |
| 5 | C ELEGANS ANAT | 33052 | 0% | 26% | 12 |
| 6 | DEV GENOM | 31680 | 0% | 33% | 9 |
| 7 | PROGRAM MOL CELLULAR DEV BIOL | 27796 | 1% | 15% | 18 |
| 8 | SCI B303 | 23768 | 0% | 75% | 3 |
| 9 | HOWARD HUGHES MED | 23545 | 10% | 1% | 289 |
| 10 | INDIANA UNIV MED SCI | 21128 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DEVELOPMENT | 110055 | 13% | 3% | 363 |
| 2 | DEVELOPMENTAL BIOLOGY | 108965 | 13% | 3% | 376 |
| 3 | GENETICS | 49940 | 9% | 2% | 246 |
| 4 | DEVELOPMENTAL CELL | 18905 | 2% | 3% | 68 |
| 5 | G3-GENES GENOMES GENETICS | 10657 | 1% | 3% | 37 |
| 6 | GENES & DEVELOPMENT | 9666 | 3% | 1% | 83 |
| 7 | CURRENT BIOLOGY | 8898 | 3% | 1% | 87 |
| 8 | WILEY INTERDISCIPLINARY REVIEWS-DEVELOPMENTAL BIOLOGY | 7221 | 0% | 6% | 12 |
| 9 | PLOS GENETICS | 6222 | 2% | 1% | 60 |
| 10 | CELL | 5851 | 3% | 1% | 92 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | C ELEGANS | 1399970 | 19% | 24% | 545 | Search C+ELEGANS | Search C+ELEGANS |
| 2 | CAENORHABDITIS ELEGANS | 460569 | 12% | 13% | 343 | Search CAENORHABDITIS+ELEGANS | Search CAENORHABDITIS+ELEGANS |
| 3 | P GRANULES | 202820 | 1% | 80% | 24 | Search P+GRANULES | Search P+GRANULES |
| 4 | VULVAL DEVELOPMENT | 186343 | 1% | 84% | 21 | Search VULVAL+DEVELOPMENT | Search VULVAL+DEVELOPMENT |
| 5 | POP 1 | 158460 | 1% | 100% | 15 | Search POP+1 | Search POP+1 |
| 6 | VULVA DEVELOPMENT | 147896 | 0% | 100% | 14 | Search VULVA+DEVELOPMENT | Search VULVA+DEVELOPMENT |
| 7 | LIN 12 | 128033 | 1% | 61% | 20 | Search LIN+12 | Search LIN+12 |
| 8 | HLH 2 | 119018 | 0% | 87% | 13 | Search HLH+2 | Search HLH+2 |
| 9 | MAB 5 | 116204 | 0% | 100% | 11 | Search MAB+5 | Search MAB+5 |
| 10 | GERMLINE | 106731 | 3% | 12% | 86 | Search GERMLINE | Search GERMLINE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ROBERTSON, S , LIN, R , (2015) THE MATERNAL-TO-ZYGOTIC TRANSITION IN C. ELEGANS.MATERNAL-TO-ZYGOTIC TRANSITION. VOL. 113. ISSUE . P. 1 -42 | 111 | 74% | 4 |
| 2 | MADURO, MF , (2010) CELL FATE SPECIFICATION IN THE C. ELEGANS EMBRYO.DEVELOPMENTAL DYNAMICS. VOL. 239. ISSUE 5. P. 1315 -1329 | 118 | 75% | 15 |
| 3 | SCHMID, T , HAJNAL, A , (2015) SIGNAL TRANSDUCTION DURING C. ELEGANS VULVAL DEVELOPMENT: A NEVER ENDING STORY.CURRENT OPINION IN GENETICS & DEVELOPMENT. VOL. 32. ISSUE . P. 1 -9 | 86 | 88% | 5 |
| 4 | GRIFFIN, EE , (2015) CYTOPLASMIC LOCALIZATION AND ASYMMETRIC DIVISION IN THE EARLY EMBRYO OF CAENORHABDITIS ELEGANS.WILEY INTERDISCIPLINARY REVIEWS-DEVELOPMENTAL BIOLOGY. VOL. 4. ISSUE 3. P. 267 -282 | 94 | 76% | 3 |
| 5 | ZACHARIAS, AL , MURRAY, JI , (2016) COMBINATORIAL DECODING OF THE INVARIANT C-ELEGANS EMBRYONIC LINEAGE IN SPACE AND TIME.GENESIS. VOL. 54. ISSUE 4. P. 182 -197 | 98 | 74% | 0 |
| 6 | WANG, JT , SEYDOUX, G , (2013) GERM CELL SPECIFICATION.GERM CELL DEVELOPMENT IN C. ELEGANS. VOL. 757. ISSUE . P. 17-39 | 80 | 84% | 11 |
| 7 | JACKSON, BM , EISENMANN, DM , (2012) BETA-CATENIN-DEPENDENT WNT SIGNALING IN C. ELEGANS: TEACHING AN OLD DOG A NEW TRICK.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 4. ISSUE 8. P. - | 89 | 74% | 12 |
| 8 | MADURO, MF , (2009) STRUCTURE AND EVOLUTION OF THE C. ELEGANS EMBRYONIC ENDOMESODERM NETWORK.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1789. ISSUE 4. P. 250 -260 | 88 | 79% | 24 |
| 9 | HANSEN, D , SCHEDL, T , (2013) STEM CELL PROLIFERATION VERSUS MEIOTIC FATE DECISION IN CAENORHABDITIS ELEGANS.GERM CELL DEVELOPMENT IN C. ELEGANS. VOL. 757. ISSUE . P. 71-99 | 76 | 74% | 21 |
| 10 | OWRAGHI, M , BROITMAN-MADURO, G , LUU, T , ROBERSON, H , MADURO, MF , (2010) ROLES OF THE WNT EFFECTOR POP-1/TCF IN THE C. ELEGANS ENDOMESODERM SPECIFICATION GENE NETWORK.DEVELOPMENTAL BIOLOGY. VOL. 340. ISSUE 2. P. 209-221 | 71 | 88% | 18 |
Classes with closest relation at Level 1 |