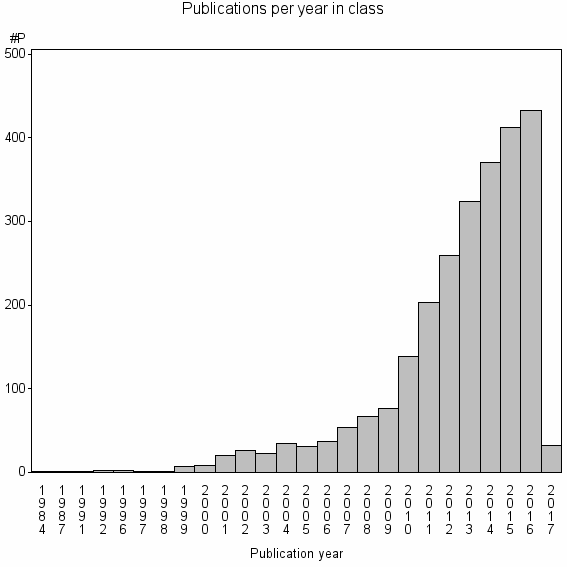
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 1631 | 2562 | 54.4 | 93% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 563 | 3 | DNA METHYLATION//EPIGENETICS//METHYLATION | 17086 |
| 382 | 2 | DNA METHYLATION//EPIGENETICS//METHYLATION | 17086 |
| 1631 | 1 | 5 HYDROXYMETHYLCYTOSINE//DNA METHYLATION//5HMC | 2562 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | 5 HYDROXYMETHYLCYTOSINE | authKW | 1265008 | 5% | 76% | 139 |
| 2 | DNA METHYLATION | authKW | 435144 | 23% | 6% | 594 |
| 3 | 5HMC | authKW | 335726 | 1% | 78% | 36 |
| 4 | TET1 | authKW | 271154 | 1% | 71% | 32 |
| 5 | TET PROTEINS | authKW | 251732 | 1% | 81% | 26 |
| 6 | DNA DEMETHYLATION | authKW | 240003 | 2% | 32% | 63 |
| 7 | TET | authKW | 238435 | 2% | 49% | 41 |
| 8 | 5 HYDROXYMETHYLCYTOSINE 5HMC | authKW | 225133 | 1% | 82% | 23 |
| 9 | 5 METHYLCYTOSINE | authKW | 172378 | 2% | 23% | 64 |
| 10 | DNA HYDROXYMETHYLATION | authKW | 165875 | 1% | 61% | 23 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 17401 | 35% | 0% | 886 |
| 2 | Cell Biology | 5944 | 26% | 0% | 658 |
| 3 | Biochemistry & Molecular Biology | 5617 | 40% | 0% | 1032 |
| 4 | Biotechnology & Applied Microbiology | 3409 | 17% | 0% | 423 |
| 5 | Developmental Biology | 2071 | 6% | 0% | 155 |
| 6 | Mathematical & Computational Biology | 1498 | 5% | 0% | 121 |
| 7 | Cell & Tissue Engineering | 1221 | 2% | 0% | 54 |
| 8 | Biochemical Research Methods | 718 | 6% | 0% | 166 |
| 9 | Oncology | 359 | 8% | 0% | 211 |
| 10 | Reproductive Biology | 179 | 2% | 0% | 51 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | EPIGENET PROGRAMME | 147428 | 1% | 38% | 33 |
| 2 | MAMMALIAN EPIGENET STUDIES | 118460 | 1% | 76% | 13 |
| 3 | EPIGENET | 68072 | 3% | 8% | 68 |
| 4 | SIGNALING GENE EXP S | 54414 | 1% | 35% | 13 |
| 5 | TROPHOBLAST | 54242 | 1% | 14% | 33 |
| 6 | BIOPHYS DYNAM | 42722 | 2% | 6% | 57 |
| 7 | GRP DNA METABSTATE MOL BIOL | 35750 | 0% | 100% | 3 |
| 8 | MAMMALIAN DEV GRP | 35750 | 0% | 100% | 3 |
| 9 | SHANDONG UNIV CUHK SDU JOINT REPROD GENET | 35750 | 0% | 100% | 3 |
| 10 | GENET COMPUTAT GENET | 34340 | 0% | 41% | 7 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | EPIGENOMICS | 59343 | 2% | 10% | 48 |
| 2 | EPIGENETICS & CHROMATIN | 34626 | 1% | 10% | 29 |
| 3 | GENOME RESEARCH | 16706 | 2% | 3% | 44 |
| 4 | EPIGENETICS | 15049 | 1% | 7% | 18 |
| 5 | CIBA FOUNDATION SYMPOSIA | 13655 | 2% | 2% | 56 |
| 6 | GENOME BIOLOGY | 12894 | 2% | 2% | 57 |
| 7 | BRIEFINGS IN FUNCTIONAL GENOMICS | 8565 | 1% | 5% | 15 |
| 8 | CELL REPORTS | 6687 | 2% | 1% | 43 |
| 9 | CLINICAL EPIGENETICS | 5726 | 1% | 3% | 14 |
| 10 | CELL STEM CELL | 5696 | 1% | 2% | 22 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | 5 HYDROXYMETHYLCYTOSINE | 1265008 | 5% | 76% | 139 | Search 5+HYDROXYMETHYLCYTOSINE | Search 5+HYDROXYMETHYLCYTOSINE |
| 2 | DNA METHYLATION | 435144 | 23% | 6% | 594 | Search DNA+METHYLATION | Search DNA+METHYLATION |
| 3 | 5HMC | 335726 | 1% | 78% | 36 | Search 5HMC | Search 5HMC |
| 4 | TET1 | 271154 | 1% | 71% | 32 | Search TET1 | Search TET1 |
| 5 | TET PROTEINS | 251732 | 1% | 81% | 26 | Search TET+PROTEINS | Search TET+PROTEINS |
| 6 | DNA DEMETHYLATION | 240003 | 2% | 32% | 63 | Search DNA+DEMETHYLATION | Search DNA+DEMETHYLATION |
| 7 | TET | 238435 | 2% | 49% | 41 | Search TET | Search TET |
| 8 | 5 HYDROXYMETHYLCYTOSINE 5HMC | 225133 | 1% | 82% | 23 | Search 5+HYDROXYMETHYLCYTOSINE+5HMC | Search 5+HYDROXYMETHYLCYTOSINE+5HMC |
| 9 | 5 METHYLCYTOSINE | 172378 | 2% | 23% | 64 | Search 5+METHYLCYTOSINE | Search 5+METHYLCYTOSINE |
| 10 | DNA HYDROXYMETHYLATION | 165875 | 1% | 61% | 23 | Search DNA+HYDROXYMETHYLATION | Search DNA+HYDROXYMETHYLATION |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | WU, H , ZHANG, Y , (2014) REVERSING DNA METHYLATION: MECHANISMS, GENOMICS, AND BIOLOGICAL FUNCTIONS.CELL. VOL. 156. ISSUE 1-2. P. 45-68 | 161 | 77% | 228 |
| 2 | RASMUSSEN, KD , HELIN, K , (2016) ROLE OF TET ENZYMES IN DNA METHYLATION, DEVELOPMENT, AND CANCER.GENES & DEVELOPMENT. VOL. 30. ISSUE 7. P. 733 -750 | 109 | 72% | 20 |
| 3 | PASTOR, WA , ARAVIND, L , RAO, A , (2013) TETONIC SHIFT: BIOLOGICAL ROLES OF TET PROTEINS IN DNA DEMETHYLATION AND TRANSCRIPTION.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 14. ISSUE 6. P. 341 -356 | 121 | 68% | 213 |
| 4 | SCHUBELER, D , (2015) FUNCTION AND INFORMATION CONTENT OF DNA METHYLATION.NATURE. VOL. 517. ISSUE 7534. P. 321 -326 | 75 | 65% | 197 |
| 5 | SMITH, ZD , MEISSNER, A , (2013) DNA METHYLATION: ROLES IN MAMMALIAN DEVELOPMENT.NATURE REVIEWS GENETICS. VOL. 14. ISSUE 3. P. 204 -220 | 100 | 51% | 500 |
| 6 | SHEN, L , SONG, CX , HE, C , ZHANG, Y , (2014) MECHANISM AND FUNCTION OF OXIDATIVE REVERSAL OF DNA AND RNA METHYLATION.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 83. VOL. 83. ISSUE . P. 585 -+ | 124 | 72% | 62 |
| 7 | KOHLI, RM , ZHANG, Y , (2013) TET ENZYMES, TDG AND THE DYNAMICS OF DNA DEMETHYLATION.NATURE. VOL. 502. ISSUE 7472. P. 472-479 | 78 | 79% | 283 |
| 8 | SCHUERMANN, D , SCHAR, P , WEBER, AR , (2016) ACTIVE DNA DEMETHYLATION BY DNA REPAIR: FACTS AND UNCERTAINTIES.DNA REPAIR. VOL. 44. ISSUE . P. 92 -102 | 118 | 82% | 2 |
| 9 | GUIBERT, S , WEBER, M , (2013) FUNCTIONS OF DNA METHYLATION AND HYDROXYMETHYLATION IN MAMMALIAN DEVELOPMENT.EPIGENETICS AND DEVELOPMENT. VOL. 104. ISSUE . P. 47 -83 | 142 | 71% | 22 |
| 10 | CADET, J , WAGNER, JR , (2014) TET ENZYMATIC OXIDATION OF 5-METHYLCYTOSINE, 5-HYDROXYMETHYLCYTOSINE AND 5-FORMYLCYTOSINE.MUTATION RESEARCH-GENETIC TOXICOLOGY AND ENVIRONMENTAL MUTAGENESIS. VOL. 764. ISSUE . P. 18-35 | 137 | 65% | 14 |
Classes with closest relation at Level 1 |