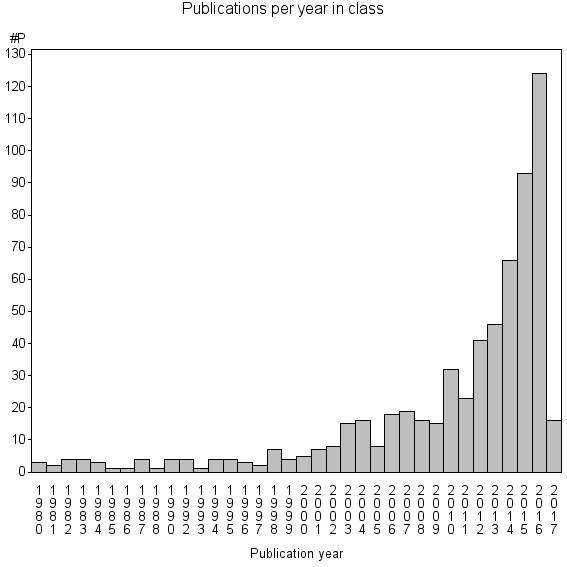
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 16988 | 624 | 47.8 | 86% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 1084 | 2 | AMINOACYL TRNA SYNTHETASE//TRNA//AMINOACYL TRANSFER RNA SYNTHETASE | 9879 |
| 16988 | 1 | DNMT2//ALKB//N 6 METHYLADENOSINE | 624 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | DNMT2 | authKW | 799240 | 3% | 78% | 21 |
| 2 | ALKB | authKW | 797460 | 6% | 44% | 37 |
| 3 | N 6 METHYLADENOSINE | authKW | 654249 | 3% | 70% | 19 |
| 4 | RNA METHYLATION | authKW | 410794 | 3% | 44% | 19 |
| 5 | N 6 METHYLADENOSINE M6A | authKW | 293602 | 1% | 100% | 6 |
| 6 | METTL3 | authKW | 266413 | 1% | 78% | 7 |
| 7 | NSUN2 | authKW | 266413 | 1% | 78% | 7 |
| 8 | ALKB HOMOLOGS | authKW | 244668 | 1% | 100% | 5 |
| 9 | ALKBH3 | authKW | 244668 | 1% | 100% | 5 |
| 10 | ALKBH2 | authKW | 203888 | 1% | 83% | 5 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 3062 | 58% | 0% | 359 |
| 2 | Cell Biology | 1178 | 23% | 0% | 146 |
| 3 | Genetics & Heredity | 657 | 15% | 0% | 91 |
| 4 | Biochemical Research Methods | 160 | 6% | 0% | 39 |
| 5 | Biophysics | 90 | 5% | 0% | 34 |
| 6 | Cell & Tissue Engineering | 78 | 1% | 0% | 7 |
| 7 | Developmental Biology | 74 | 3% | 0% | 16 |
| 8 | Toxicology | 59 | 4% | 0% | 23 |
| 9 | Biotechnology & Applied Microbiology | 37 | 5% | 0% | 30 |
| 10 | Parasitiology | 27 | 2% | 0% | 10 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOPHYS DYNAM | 124625 | 8% | 5% | 48 |
| 2 | BIG CAS OSLO GENOME COOPERAT | 97867 | 0% | 100% | 2 |
| 3 | ENGN SPACE BIOL | 97867 | 0% | 100% | 2 |
| 4 | HRINU | 97867 | 0% | 100% | 2 |
| 5 | MOL CELLULAR BIOCHEM BBSRB | 97867 | 0% | 100% | 2 |
| 6 | SUERI | 97867 | 0% | 100% | 2 |
| 7 | XJTLU WTNC | 97867 | 0% | 100% | 2 |
| 8 | CLIN DIAGNOST INTERVENT | 83878 | 1% | 29% | 6 |
| 9 | UBIQUITIN SIGNALLING GRP | 73397 | 0% | 50% | 3 |
| 10 | MICROBIOL DIAGNOST INTERVENT | 65243 | 0% | 67% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 6136 | 3% | 1% | 16 |
| 2 | NUCLEIC ACIDS RESEARCH | 2803 | 8% | 0% | 48 |
| 3 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 2125 | 1% | 1% | 4 |
| 4 | NATURE CHEMICAL BIOLOGY | 1738 | 1% | 1% | 7 |
| 5 | CELL CHEMICAL BIOLOGY | 1394 | 0% | 1% | 2 |
| 6 | RNA BIOLOGY | 1324 | 1% | 1% | 5 |
| 7 | RNA | 933 | 1% | 0% | 7 |
| 8 | NATURE STRUCTURAL & MOLECULAR BIOLOGY | 804 | 1% | 0% | 6 |
| 9 | JOURNAL OF MOLECULAR CELL BIOLOGY | 735 | 0% | 1% | 2 |
| 10 | CELL | 713 | 2% | 0% | 15 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNMT2 | 799240 | 3% | 78% | 21 | Search DNMT2 | Search DNMT2 |
| 2 | ALKB | 797460 | 6% | 44% | 37 | Search ALKB | Search ALKB |
| 3 | N 6 METHYLADENOSINE | 654249 | 3% | 70% | 19 | Search N+6+METHYLADENOSINE | Search N+6+METHYLADENOSINE |
| 4 | RNA METHYLATION | 410794 | 3% | 44% | 19 | Search RNA+METHYLATION | Search RNA+METHYLATION |
| 5 | N 6 METHYLADENOSINE M6A | 293602 | 1% | 100% | 6 | Search N+6+METHYLADENOSINE+M6A | Search N+6+METHYLADENOSINE+M6A |
| 6 | METTL3 | 266413 | 1% | 78% | 7 | Search METTL3 | Search METTL3 |
| 7 | NSUN2 | 266413 | 1% | 78% | 7 | Search NSUN2 | Search NSUN2 |
| 8 | ALKB HOMOLOGS | 244668 | 1% | 100% | 5 | Search ALKB+HOMOLOGS | Search ALKB+HOMOLOGS |
| 9 | ALKBH3 | 244668 | 1% | 100% | 5 | Search ALKBH3 | Search ALKBH3 |
| 10 | ALKBH2 | 203888 | 1% | 83% | 5 | Search ALKBH2 | Search ALKBH2 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ZHAO, BXS , ROUNDTREE, IA , HE, C , (2017) POST-TRANSCRIPTIONAL GENE REGULATION BY MRNA MODIFICATIONS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 18. ISSUE 1. P. 31 -42 | 87 | 59% | 1 |
| 2 | ZHENG, GQ , FU, Y , HE, C , (2014) NUCLEIC ACID OXIDATION IN DNA DAMAGE REPAIR AND EPIGENETICS.CHEMICAL REVIEWS. VOL. 114. ISSUE 8. P. 4602 -4620 | 111 | 53% | 19 |
| 3 | ROUNDTREE, IA , HE, C , (2016) RNA EPIGENETICS - CHEMICAL MESSAGES FOR POSTTRANSCRIPTIONAL GENE REGULATION.CURRENT OPINION IN CHEMICAL BIOLOGY. VOL. 30. ISSUE . P. 46 -51 | 40 | 93% | 11 |
| 4 | FEDELES, BI , SINGH, V , DELANEY, JC , LI, DY , ESSIGMANN, JM , (2015) THE ALKB FAMILY OF FE(II)/ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASES: REPAIRING NUCLEIC ACID ALKYLATION DAMAGE AND BEYOND.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 290. ISSUE 34. P. 20734 -20742 | 65 | 72% | 10 |
| 5 | OUGLAND, R , ROGNES, T , KLUNGLAND, A , LARSEN, E , (2015) NON-HOMOLOGOUS FUNCTIONS OF THE ALKB HOMOLOGS.JOURNAL OF MOLECULAR CELL BIOLOGY. VOL. 7. ISSUE 6. P. 494 -504 | 75 | 64% | 3 |
| 6 | LIN, SB , CHOE, J , DU, P , TRIBOULET, R , GREGORY, RI , (2016) THE M(6)A METHYLTRANSFERASE METTL3 PROMOTES TRANSLATION IN HUMAN CANCER CELLS.MOLECULAR CELL. VOL. 62. ISSUE 3. P. 335 -345 | 32 | 82% | 11 |
| 7 | WANG, Y , ZHAO, JC , (2016) UPDATE: MECHANISMS UNDERLYING N-6-METHYLADENOSINE MODIFICATION OF EUKARYOTIC MRNA.TRENDS IN GENETICS. VOL. 32. ISSUE 12. P. 763 -773 | 54 | 79% | 0 |
| 8 | MEYER, KD , JAFFREY, SR , (2014) THE DYNAMIC EPITRANSCRIPTOME: N-6-METHYLADENOSINE AND GENE EXPRESSION CONTROL.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 15. ISSUE 5. P. 313 -326 | 48 | 53% | 102 |
| 9 | YUE, YN , LIU, JZ , HE, C , (2015) RNA N-6-METHYLADENOSINE METHYLATION IN POST-TRANSCRIPTIONAL GENE EXPRESSION REGULATION.GENES & DEVELOPMENT. VOL. 29. ISSUE 13. P. 1343 -1355 | 53 | 55% | 30 |
| 10 | SCHWARTZ, S , (2016) CRACKING THE EPITRANSCRIPTOME.RNA. VOL. 22. ISSUE 2. P. 169 -174 | 39 | 72% | 7 |
Classes with closest relation at Level 1 |