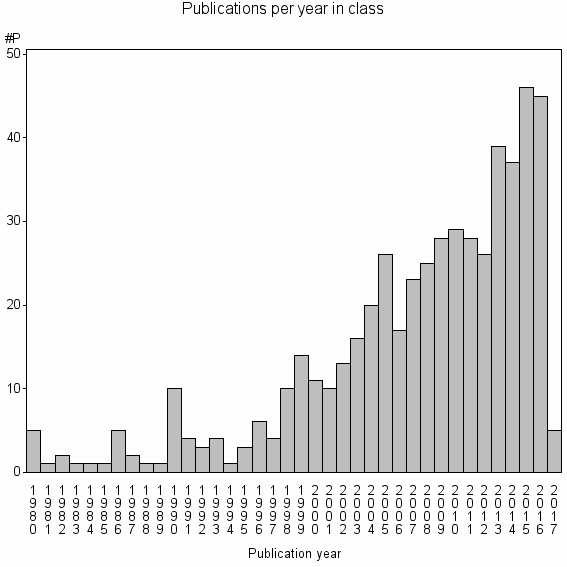
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 19084 | 523 | 44.3 | 88% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 1084 | 2 | AMINOACYL TRNA SYNTHETASE//TRNA//AMINOACYL TRANSFER RNA SYNTHETASE | 9879 |
| 19084 | 1 | TRNASE Z//TRNA NUCLEOTIDYLTRANSFERASE//CCA ADDING ENZYME | 523 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | TRNASE Z | authKW | 1112072 | 4% | 95% | 20 |
| 2 | TRNA NUCLEOTIDYLTRANSFERASE | authKW | 560481 | 2% | 80% | 12 |
| 3 | CCA ADDING ENZYME | authKW | 525449 | 2% | 75% | 12 |
| 4 | TRNA DERIVED FRAGMENTS | authKW | 449102 | 2% | 77% | 10 |
| 5 | TRNASE ZL | authKW | 415173 | 2% | 89% | 8 |
| 6 | TRUE GENE SILENCING | authKW | 408687 | 1% | 100% | 7 |
| 7 | 3 TRNASE | authKW | 357600 | 1% | 88% | 7 |
| 8 | TRNA 3 PROCESSING | authKW | 350303 | 1% | 100% | 6 |
| 9 | NUCLEAR TRNA EXPORT | authKW | 300258 | 1% | 86% | 6 |
| 10 | TRNA FRAGMENT | authKW | 291920 | 1% | 100% | 5 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 4374 | 73% | 0% | 384 |
| 2 | Cell Biology | 1148 | 25% | 0% | 131 |
| 3 | Genetics & Heredity | 306 | 11% | 0% | 59 |
| 4 | Biophysics | 175 | 8% | 0% | 40 |
| 5 | Developmental Biology | 94 | 3% | 0% | 16 |
| 6 | Mycology | 45 | 1% | 0% | 7 |
| 7 | Biotechnology & Applied Microbiology | 27 | 5% | 0% | 24 |
| 8 | Biology | 25 | 3% | 0% | 14 |
| 9 | Parasitiology | 20 | 2% | 0% | 8 |
| 10 | Biochemical Research Methods | 16 | 3% | 0% | 15 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JIANGSU MICROBES GENOM | 131362 | 1% | 75% | 3 |
| 2 | RNA BIOL | 97564 | 7% | 5% | 36 |
| 3 | AKIHAKU | 58384 | 0% | 100% | 1 |
| 4 | BOT 3038 HAWORTH HALL | 58384 | 0% | 100% | 1 |
| 5 | CANC GENOM PROT | 58384 | 0% | 100% | 1 |
| 6 | FUNCT GENOM DISCOVERY TECHNOL | 58384 | 0% | 100% | 1 |
| 7 | IBIMED HLTH SCI | 58384 | 0% | 100% | 1 |
| 8 | INSERM UMR773 GENET | 58384 | 0% | 100% | 1 |
| 9 | MAX PLANCK EVOLUT ANTHROPOL | 58384 | 0% | 100% | 1 |
| 10 | MED ORLANDO | 58384 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA | 18030 | 5% | 1% | 28 |
| 2 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 5715 | 1% | 2% | 6 |
| 3 | RNA BIOLOGY | 5139 | 2% | 1% | 9 |
| 4 | NUCLEIC ACIDS RESEARCH | 4269 | 10% | 0% | 54 |
| 5 | RNA-A PUBLICATION OF THE RNA SOCIETY | 2237 | 1% | 1% | 7 |
| 6 | FRONTIERS IN GENETICS | 1161 | 1% | 1% | 4 |
| 7 | JOURNAL OF BIOLOGICAL CHEMISTRY | 982 | 10% | 0% | 52 |
| 8 | GENES & DEVELOPMENT | 932 | 2% | 0% | 11 |
| 9 | NUCLEUS | 871 | 0% | 1% | 2 |
| 10 | MOLECULAR BIOLOGY OF THE CELL | 685 | 2% | 0% | 10 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TRNASE Z | 1112072 | 4% | 95% | 20 | Search TRNASE+Z | Search TRNASE+Z |
| 2 | TRNA NUCLEOTIDYLTRANSFERASE | 560481 | 2% | 80% | 12 | Search TRNA+NUCLEOTIDYLTRANSFERASE | Search TRNA+NUCLEOTIDYLTRANSFERASE |
| 3 | CCA ADDING ENZYME | 525449 | 2% | 75% | 12 | Search CCA+ADDING+ENZYME | Search CCA+ADDING+ENZYME |
| 4 | TRNA DERIVED FRAGMENTS | 449102 | 2% | 77% | 10 | Search TRNA+DERIVED+FRAGMENTS | Search TRNA+DERIVED+FRAGMENTS |
| 5 | TRNASE ZL | 415173 | 2% | 89% | 8 | Search TRNASE+ZL | Search TRNASE+ZL |
| 6 | TRUE GENE SILENCING | 408687 | 1% | 100% | 7 | Search TRUE+GENE+SILENCING | Search TRUE+GENE+SILENCING |
| 7 | 3 TRNASE | 357600 | 1% | 88% | 7 | Search 3+TRNASE | Search 3+TRNASE |
| 8 | TRNA 3 PROCESSING | 350303 | 1% | 100% | 6 | Search TRNA+3+PROCESSING | Search TRNA+3+PROCESSING |
| 9 | NUCLEAR TRNA EXPORT | 300258 | 1% | 86% | 6 | Search NUCLEAR+TRNA+EXPORT | Search NUCLEAR+TRNA+EXPORT |
| 10 | TRNA FRAGMENT | 291920 | 1% | 100% | 5 | Search TRNA+FRAGMENT | Search TRNA+FRAGMENT |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | PHIZICKY, EM , HOPPER, AK , (2010) TRNA BIOLOGY CHARGES TO THE FRONT.GENES & DEVELOPMENT. VOL. 24. ISSUE 17. P. 1832-1860 | 91 | 25% | 228 |
| 2 | LOHER, P , TELONIS, AG , RIGOUTSOS, I , (2017) MINTMAP: FAST AND EXHAUSTIVE PROFILING OF NUCLEAR AND MITOCHONDRIAL TRNA FRAGMENTS FROM SHORT RNA-SEQ DATA.SCIENTIFIC REPORTS. VOL. 7. ISSUE . P. 1 -20 | 42 | 71% | 0 |
| 3 | MEGEL, C , MORELLE, G , LALANDE, S , DUCHENE, AM , SMALL, I , MARECHAL-DROUARD, L , (2015) SURVEILLANCE AND CLEAVAGE OF EUKARYOTIC TRNAS.INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. VOL. 16. ISSUE 1. P. 1873 -1893 | 50 | 52% | 8 |
| 4 | WILUSZ, JE , (2015) CONTROLLING TRANSLATION VIA MODULATION OF TRNA LEVELS.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 6. ISSUE 4. P. 453 -470 | 59 | 43% | 2 |
| 5 | FAN, LJ , WANG, ZK , LIU, JY , GUO, WL , YAN, J , HUANG, Y , (2011) A SURVEY OF GREEN PLANT TRNA 3 '-END PROCESSING ENZYME TRNASE ZS, HOMOLOGS OF THE CANDIDATE PROSTATE CANCER SUSCEPTIBILITY PROTEIN ELAC2.BMC EVOLUTIONARY BIOLOGY. VOL. 11. ISSUE . P. - | 45 | 61% | 4 |
| 6 | LEVINGER, L , HOPKINSON, A , DESETTY, R , WILSON, C , (2009) EFFECT OF CHANGES IN THE FLEXIBLE ARM ON TRNASE Z PROCESSING KINETICS.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 284. ISSUE 23. P. 15685-15691 | 31 | 86% | 3 |
| 7 | TOMITA, K , YAMASHITA, S , (2014) MOLECULAR MECHANISMS OF TEMPLATE-INDEPENDENT RNA POLYMERIZATION BY TRNA NUCLEOTIDYLTRANSFERASES.FRONTIERS IN GENETICS. VOL. 5. ISSUE . P. - | 30 | 73% | 8 |
| 8 | CEBALLOS, M , VIOQUE, A , (2007) TRNASE Z.PROTEIN AND PEPTIDE LETTERS. VOL. 14. ISSUE 2. P. 137-145 | 44 | 60% | 13 |
| 9 | GEBETSBERGER, J , POLACEK, N , (2013) SLICING TRNAS TO BOOST FUNCTIONAL NCRNA DIVERSITY.RNA BIOLOGY. VOL. 10. ISSUE 12. P. 1798-1806 | 38 | 48% | 48 |
| 10 | HAINO, A , ISHIKAWA, T , SEKI, M , NASHIMOTO, M , (2016) TRUE GENE SILENCING: SCREENING OF A HEPTAMER-TYPE SMALL GUIDE RNA LIBRARY FOR POTENTIAL CANCER THERAPEUTIC AGENTS.JOVE-JOURNAL OF VISUALIZED EXPERIMENTS. VOL. . ISSUE 112. P. - | 21 | 100% | 0 |
Classes with closest relation at Level 1 |