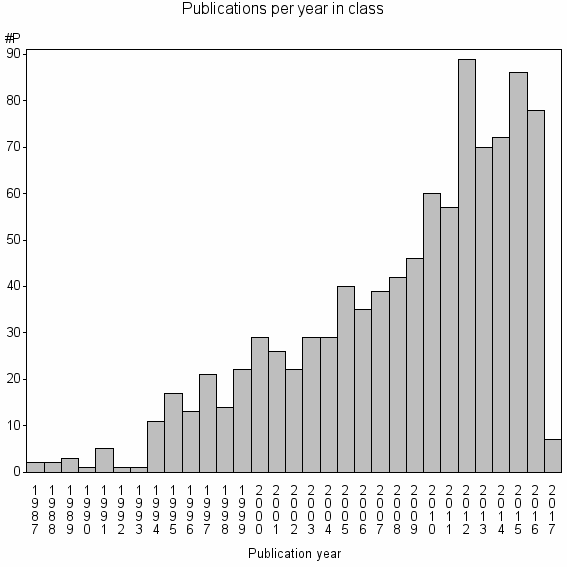
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 11597 | 969 | 49.3 | 93% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 227 | 3 | MICRORNA//MIRNA//GENE DELIVERY | 49432 |
| 3423 | 2 | RNA EDITING//ADAR//AEG 1 | 1570 |
| 11597 | 1 | RNA EDITING//ADAR//ADAR1 | 969 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RNA EDITING | authKW | 2344198 | 28% | 27% | 274 |
| 2 | ADAR | authKW | 2341591 | 9% | 83% | 90 |
| 3 | ADAR1 | authKW | 952493 | 5% | 66% | 46 |
| 4 | ADAR2 | authKW | 729411 | 3% | 93% | 25 |
| 5 | ADARS | authKW | 378129 | 1% | 100% | 12 |
| 6 | A TO I RNA EDITING | authKW | 366661 | 2% | 73% | 16 |
| 7 | SEROTONIN 2C RECEPTOR | authKW | 315965 | 2% | 53% | 19 |
| 8 | ADENOSINE DEAMINATION | authKW | 313248 | 1% | 76% | 13 |
| 9 | ADENOSINE DEAMINASE ACTING ON RNA ADAR | authKW | 220575 | 1% | 100% | 7 |
| 10 | INOSINE | authKW | 191451 | 5% | 12% | 49 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 3390 | 49% | 0% | 479 |
| 2 | Cell Biology | 1186 | 19% | 0% | 187 |
| 3 | Genetics & Heredity | 1015 | 15% | 0% | 141 |
| 4 | Neurosciences | 650 | 17% | 0% | 166 |
| 5 | Virology | 246 | 4% | 0% | 42 |
| 6 | Biotechnology & Applied Microbiology | 216 | 8% | 0% | 75 |
| 7 | Biochemical Research Methods | 164 | 5% | 0% | 51 |
| 8 | Mathematical & Computational Biology | 44 | 2% | 0% | 15 |
| 9 | Developmental Biology | 22 | 1% | 0% | 13 |
| 10 | Biophysics | 22 | 3% | 0% | 28 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA EDITING | 378129 | 1% | 100% | 12 |
| 2 | MOL BIOL FUNCT GENOM | 55004 | 2% | 9% | 20 |
| 3 | CHROMOSOME BIOL | 46819 | 2% | 7% | 20 |
| 4 | INTER PROGRAM BIOCHEM MOL BIOL | 46332 | 1% | 29% | 5 |
| 5 | POLE EXPT AVICOLE TOURS | 42013 | 0% | 67% | 2 |
| 6 | ANIM STEM CELL PROJECT | 31511 | 0% | 100% | 1 |
| 7 | BERKELEY DROSOPHILA GENOME PROJECTLIFE SCI | 31511 | 0% | 100% | 1 |
| 8 | BIOCHEM 50 N MED DR | 31511 | 0% | 100% | 1 |
| 9 | BIOL COMPLEX SYST MODELING ENGN DIAG | 31511 | 0% | 100% | 1 |
| 10 | BIOMED ENGN MOL BIOPHYSMINIST EDUC | 31511 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA BIOLOGY | 11098 | 2% | 2% | 18 |
| 2 | RNA-A PUBLICATION OF THE RNA SOCIETY | 10892 | 2% | 2% | 21 |
| 3 | RNA | 7732 | 3% | 1% | 25 |
| 4 | GENOME RESEARCH | 4467 | 1% | 1% | 14 |
| 5 | NUCLEIC ACIDS RESEARCH | 1848 | 5% | 0% | 49 |
| 6 | GENOME BIOLOGY | 1763 | 1% | 0% | 13 |
| 7 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 1366 | 0% | 1% | 4 |
| 8 | NEUROSCIENCE RESEARCH | 1244 | 1% | 0% | 12 |
| 9 | CURRENT ISSUES IN MOLECULAR BIOLOGY | 930 | 0% | 1% | 2 |
| 10 | NATURE STRUCTURAL & MOLECULAR BIOLOGY | 919 | 1% | 0% | 8 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA EDITING | 2344198 | 28% | 27% | 274 | Search RNA+EDITING | Search RNA+EDITING |
| 2 | ADAR | 2341591 | 9% | 83% | 90 | Search ADAR | Search ADAR |
| 3 | ADAR1 | 952493 | 5% | 66% | 46 | Search ADAR1 | Search ADAR1 |
| 4 | ADAR2 | 729411 | 3% | 93% | 25 | Search ADAR2 | Search ADAR2 |
| 5 | ADARS | 378129 | 1% | 100% | 12 | Search ADARS | Search ADARS |
| 6 | A TO I RNA EDITING | 366661 | 2% | 73% | 16 | Search A+TO+I+RNA+EDITING | Search A+TO+I+RNA+EDITING |
| 7 | SEROTONIN 2C RECEPTOR | 315965 | 2% | 53% | 19 | Search SEROTONIN+2C+RECEPTOR | Search SEROTONIN+2C+RECEPTOR |
| 8 | ADENOSINE DEAMINATION | 313248 | 1% | 76% | 13 | Search ADENOSINE+DEAMINATION | Search ADENOSINE+DEAMINATION |
| 9 | ADENOSINE DEAMINASE ACTING ON RNA ADAR | 220575 | 1% | 100% | 7 | Search ADENOSINE+DEAMINASE+ACTING+ON+RNA+ADAR | Search ADENOSINE+DEAMINASE+ACTING+ON+RNA+ADAR |
| 10 | INOSINE | 191451 | 5% | 12% | 49 | Search INOSINE | Search INOSINE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | NISHIKURA, K , (2016) A-TO-I EDITING OF CODING AND NON-CODING RNAS BY ADARS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 17. ISSUE 2. P. 83 -96 | 116 | 81% | 18 |
| 2 | NISHIKURA, K , (2010) FUNCTIONS AND REGULATION OF RNA EDITING BY ADAR DEAMINASES.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 79. VOL. 79. ISSUE . P. 321-349 | 120 | 80% | 343 |
| 3 | DEFFIT, SN , HUNDLEY, HA , (2016) TO EDIT OR NOT TO EDIT: REGULATION OF ADAR EDITING SPECIFICITY AND EFFICIENCY.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 7. ISSUE 1. P. 113 -127 | 108 | 90% | 1 |
| 4 | GEORGE, CX , GAN, ZJ , LIU, Y , SAMUEL, CE , (2011) ADENOSINE DEAMINASES ACTING ON RNA, RNA EDITING, AND INTERFERON ACTION.JOURNAL OF INTERFERON AND CYTOKINE RESEARCH. VOL. 31. ISSUE 1. P. 99 -117 | 132 | 69% | 36 |
| 5 | MAAS, S , (2012) POSTTRANSCRIPTIONAL RECODING BY RNA EDITING.ADVANCES IN PROTEIN CHEMISTRY AND STRUCTURAL BIOLOGY, VOL 86. VOL. 86. ISSUE . P. 193-224 | 107 | 81% | 17 |
| 6 | GALLO, A , LOCATELLI, F , (2012) ADARS: ALLIES OR ENEMIES? THE IMPORTANCE OF A-TO-I RNA EDITING IN HUMAN DISEASE: FROM CANCER TO HIV-1.BIOLOGICAL REVIEWS. VOL. 87. ISSUE 1. P. 95 -110 | 111 | 71% | 17 |
| 7 | ROSENTHAL, JJC , (2015) THE EMERGING ROLE OF RNA EDITING IN PLASTICITY.JOURNAL OF EXPERIMENTAL BIOLOGY. VOL. 218. ISSUE 12. P. 1812 -1821 | 87 | 76% | 9 |
| 8 | SAMUEL, CE , (2011) ADENOSINE DEAMINASES ACTING ON RNA (ADARS) ARE BOTH ANTIVIRAL AND PROVIRAL.VIROLOGY. VOL. 411. ISSUE 2. P. 180 -193 | 110 | 60% | 73 |
| 9 | TAJADDOD, M , JANTSCH, MF , LICHT, K , (2016) THE DYNAMIC EPITRANSCRIPTOME: A TO I EDITING MODULATES GENETIC INFORMATION.CHROMOSOMA. VOL. 125. ISSUE 1. P. 51 -63 | 94 | 78% | 0 |
| 10 | SONG, C , SAKURAI, M , SHIROMOTO, Y , NISHIKURA, K , (2016) FUNCTIONS OF THE RNA EDITING ENZYME ADAR1 AND THEIR RELEVANCE TO HUMAN DISEASES.GENES. VOL. 7. ISSUE 12. P. - | 98 | 71% | 0 |
Classes with closest relation at Level 1 |