Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
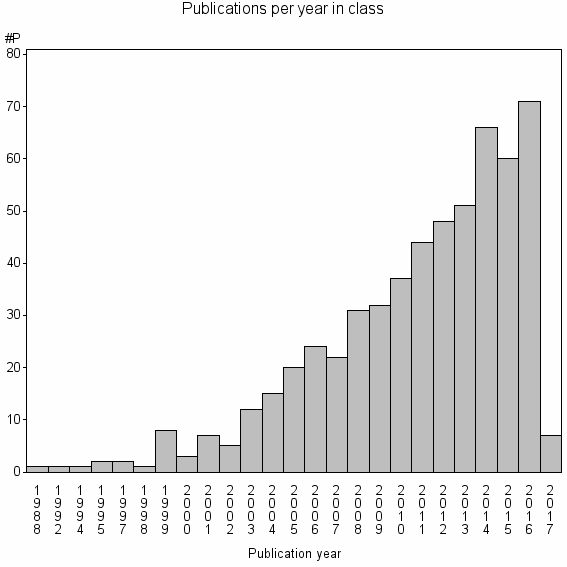
| 18004 | 571 | 48.1 | 89% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 365 | 2 | PROTEIN STRUCTURE PREDICTION//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PSEUDO AMINO ACID COMPOSITION | 17331 |
| 18004 | 1 | INTERACTION PROPENSITY//PROTEIN RNA INTERACTIONS//DNA BINDING RESIDUES | 571 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | INTERACTION PROPENSITY | authKW | 320855 | 1% | 100% | 6 |
| 2 | PROTEIN RNA INTERACTIONS | authKW | 181241 | 4% | 17% | 20 |
| 3 | DNA BINDING RESIDUES | authKW | 160427 | 1% | 100% | 3 |
| 4 | RNA BINDING RESIDUES | authKW | 160427 | 1% | 100% | 3 |
| 5 | CROSS LINKING AND IMMUNOPRECIPITATION | authKW | 106952 | 0% | 100% | 2 |
| 6 | DIRECT AND INDIRECT READOUTS | authKW | 106952 | 0% | 100% | 2 |
| 7 | DNA PROTEIN RECOGNITION MODULE | authKW | 106952 | 0% | 100% | 2 |
| 8 | INTERACTOME CAPTURE | authKW | 106952 | 0% | 100% | 2 |
| 9 | INTERFACE CLASS | authKW | 106952 | 0% | 100% | 2 |
| 10 | OFF INFORMATIZAT MANAGEMENT PLANNING | address | 106952 | 0% | 100% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 5214 | 18% | 0% | 101 |
| 2 | Biochemistry & Molecular Biology | 2699 | 57% | 0% | 323 |
| 3 | Biochemical Research Methods | 2165 | 21% | 0% | 120 |
| 4 | Biotechnology & Applied Microbiology | 1291 | 21% | 0% | 120 |
| 5 | Biophysics | 530 | 12% | 0% | 68 |
| 6 | Genetics & Heredity | 250 | 10% | 0% | 57 |
| 7 | Computer Science, Interdisciplinary Applications | 169 | 5% | 0% | 30 |
| 8 | Cell Biology | 162 | 11% | 0% | 60 |
| 9 | Biology | 63 | 4% | 0% | 21 |
| 10 | Medical Informatics | 5 | 1% | 0% | 3 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | OFF INFORMATIZAT MANAGEMENT PLANNING | 106952 | 0% | 100% | 2 |
| 2 | HLTH INFORMAT TRANSLAT SCI HITS 5021 | 71300 | 0% | 67% | 2 |
| 3 | COMPUTAT INTELLIGENCE LEARNING DISCOVERY | 60157 | 1% | 38% | 3 |
| 4 | GOLDEN AUDIT | 60157 | 1% | 38% | 3 |
| 5 | BIOCHEM MOL PHARMACOL MED | 53476 | 0% | 100% | 1 |
| 6 | BIOHEALTH INFORMAT | 53476 | 0% | 100% | 1 |
| 7 | BIOL 68 580 | 53476 | 0% | 100% | 1 |
| 8 | COLUMBIA UNIV COMPUTAT BIOL BIOINFORMAT C2B | 53476 | 0% | 100% | 1 |
| 9 | COMPUTAC BIOL | 53476 | 0% | 100% | 1 |
| 10 | COMPUTAT BIOL INFECT GRP | 53476 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NUCLEIC ACIDS RESEARCH | 13823 | 18% | 0% | 101 |
| 2 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 8022 | 5% | 0% | 31 |
| 3 | BIOINFORMATICS | 6470 | 6% | 0% | 36 |
| 4 | BMC BIOINFORMATICS | 5161 | 5% | 0% | 27 |
| 5 | GENOME BIOLOGY | 5158 | 3% | 1% | 17 |
| 6 | BRIEFINGS IN FUNCTIONAL GENOMICS | 4275 | 1% | 2% | 5 |
| 7 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 3633 | 1% | 1% | 5 |
| 8 | RNA BIOLOGY | 2844 | 1% | 1% | 7 |
| 9 | PROTEOME SCIENCE | 2399 | 1% | 1% | 5 |
| 10 | COMPUTATIONAL BIOLOGY AND CHEMISTRY | 2188 | 1% | 1% | 6 |
Author Key Words |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | YAN, J , FRIEDRICH, S , KURGAN, L , (2016) A COMPREHENSIVE COMPARATIVE REVIEW OF SEQUENCE-BASED PREDICTORS OF DNA- AND RNA-BINDING RESIDUES.BRIEFINGS IN BIOINFORMATICS. VOL. 17. ISSUE 1. P. 88 -105 | 41 | 76% | 6 |
| 2 | MIAO, Z , WESTHOF, E , (2015) A LARGE-SCALE ASSESSMENT OF NUCLEIC ACIDS BINDING SITE PREDICTION PROGRAMS.PLOS COMPUTATIONAL BIOLOGY. VOL. 11. ISSUE 12. P. - | 48 | 84% | 5 |
| 3 | MIAO, ZC , WESTHOF, E , (2015) PREDICTION OF NUCLEIC ACID BINDING PROBABILITY IN PROTEINS: A NEIGHBORING RESIDUE NETWORK BASED SCORE.NUCLEIC ACIDS RESEARCH. VOL. 43. ISSUE 11. P. 5340 -5351 | 42 | 70% | 8 |
| 4 | YANG, XX , WANG, J , SUN, J , LIU, R , (2015) SNBRFINDER: A SEQUENCE-BASED HYBRID ALGORITHM FOR ENHANCED PREDICTION OF NUCLEIC ACID-BINDING RESIDUES.PLOS ONE. VOL. 10. ISSUE 7. P. - | 43 | 73% | 1 |
| 5 | GROMIHA, MM , NAGARAJAN, R , (2013) COMPUTATIONAL APPROACHES FOR PREDICTING THE BINDING SITES AND UNDERSTANDING THE RECOGNITION MECHANISM OF PROTEIN-DNA COMPLEXES.PROTEIN-NUCLEIC ACIDS INTERACTIONS. VOL. 91. ISSUE . P. 65-99 | 55 | 55% | 4 |
| 6 | DEY, S , PAL, A , GUHAROY, M , SONAVANE, S , CHAKRABARTI, P , (2012) CHARACTERIZATION AND PREDICTION OF THE BINDING SITE IN DNA-BINDING PROTEINS: IMPROVEMENT OF ACCURACY BY COMBINING RESIDUE COMPOSITION, EVOLUTIONARY CONSERVATION AND STRUCTURAL PARAMETERS.NUCLEIC ACIDS RESEARCH. VOL. 40. ISSUE 15. P. 7150 -7161 | 39 | 75% | 9 |
| 7 | SI, JN , ZHAO, R , WU, RL , (2015) AN OVERVIEW OF THE PREDICTION OF PROTEIN DNA-BINDING SITES.INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. VOL. 16. ISSUE 3. P. 5194 -5215 | 47 | 50% | 5 |
| 8 | XIONG, Y , XIA, JF , ZHANG, W , LIU, J , (2011) EXPLOITING A REDUCED SET OF WEIGHTED AVERAGE FEATURES TO IMPROVE PREDICTION OF DNA-BINDING RESIDUES FROM 3D STRUCTURES.PLOS ONE. VOL. 6. ISSUE 12. P. - | 37 | 73% | 6 |
| 9 | BARIK, A , MISHRA, A , BAHADUR, RP , (2012) PRINCE: A WEB SERVER FOR STRUCTURAL AND PHYSICOCHEMICAL ANALYSIS OF PROTEIN-RNA INTERFACE.NUCLEIC ACIDS RESEARCH. VOL. 40. ISSUE W1. P. W440-W444 | 32 | 80% | 4 |
| 10 | YANG, XX , DENG, ZL , LIU, R , (2014) RBRDETECTOR: IMPROVED PREDICTION OF BINDING RESIDUES ON RNA-BINDING PROTEIN STRUCTURES USING COMPLEMENTARY FEATURE- AND TEMPLATE-BASED STRATEGIES.PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS. VOL. 82. ISSUE 10. P. 2455 -2471 | 31 | 74% | 6 |
Classes with closest relation at Level 1 |