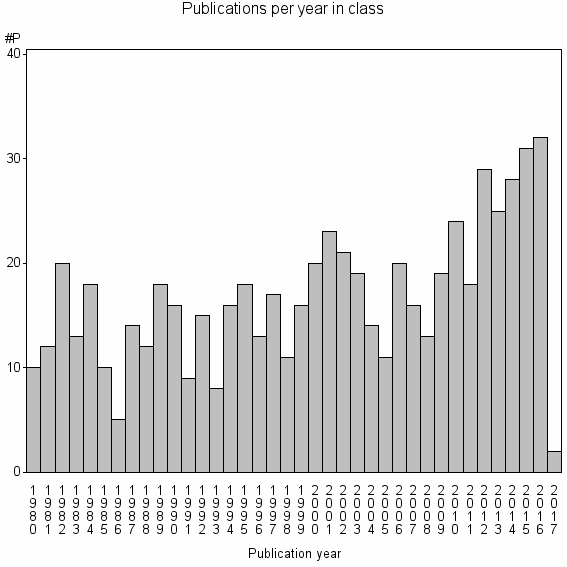
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 16776 | 636 | 46.5 | 74% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 2025 | 2 | FTSZ//PEPTIDOGLYCAN//BACTERIAL CELL DIVISION | 5424 |
| 16776 | 1 | CAULOBACTER//CTRA//CAULOBACTER CRESCENTUS | 636 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CAULOBACTER | authKW | 1455435 | 8% | 63% | 48 |
| 2 | CTRA | authKW | 660707 | 3% | 81% | 17 |
| 3 | CAULOBACTER CRESCENTUS | authKW | 550307 | 5% | 35% | 33 |
| 4 | CCRM | authKW | 216043 | 1% | 75% | 6 |
| 5 | MED BECKMAN DEV BIOL | address | 108022 | 0% | 75% | 3 |
| 6 | GCRA | authKW | 100016 | 1% | 42% | 5 |
| 7 | BIOCHEM MOL GENET BACTERIA | address | 96021 | 0% | 100% | 2 |
| 8 | CTRA RESPONSE REGULATOR | authKW | 96021 | 0% | 100% | 2 |
| 9 | PHICBK | authKW | 96021 | 0% | 100% | 2 |
| 10 | GENET GENOM GENEVA IGE3 | address | 77419 | 2% | 16% | 10 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Microbiology | 13349 | 60% | 0% | 381 |
| 2 | Biochemistry & Molecular Biology | 1379 | 40% | 0% | 255 |
| 3 | Cell Biology | 543 | 17% | 0% | 105 |
| 4 | Genetics & Heredity | 254 | 10% | 0% | 61 |
| 5 | Developmental Biology | 136 | 3% | 0% | 21 |
| 6 | Biology | 41 | 3% | 0% | 19 |
| 7 | Biochemical Research Methods | 35 | 3% | 0% | 22 |
| 8 | Mathematical & Computational Biology | 30 | 2% | 0% | 10 |
| 9 | Biotechnology & Applied Microbiology | 23 | 4% | 0% | 26 |
| 10 | Virology | 6 | 1% | 0% | 8 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MED BECKMAN DEV BIOL | 108022 | 0% | 75% | 3 |
| 2 | BIOCHEM MOL GENET BACTERIA | 96021 | 0% | 100% | 2 |
| 3 | GENET GENOM GENEVA IGE3 | 77419 | 2% | 16% | 10 |
| 4 | INTERDISCIPLINARY USR3078 | 64012 | 0% | 67% | 2 |
| 5 | QUARTIER UNIL SORGE | 54855 | 1% | 14% | 8 |
| 6 | MED CMU | 51205 | 1% | 27% | 4 |
| 7 | MICROBIOL MOL MED | 51141 | 3% | 5% | 22 |
| 8 | BECKMAN 300 | 48010 | 0% | 100% | 1 |
| 9 | BECKMAN B351 | 48010 | 0% | 100% | 1 |
| 10 | BECMKAN B300 | 48010 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF BACTERIOLOGY | 39627 | 26% | 1% | 165 |
| 2 | MOLECULAR MICROBIOLOGY | 12648 | 9% | 0% | 56 |
| 3 | CURRENT OPINION IN MICROBIOLOGY | 9459 | 3% | 1% | 19 |
| 4 | JOURNAL OF MOLECULAR BIOLOGY | 1816 | 5% | 0% | 30 |
| 5 | TRIBOLOGY & LUBRICATION TECHNOLOGY | 1249 | 1% | 1% | 4 |
| 6 | GENES & DEVELOPMENT | 1070 | 2% | 0% | 13 |
| 7 | PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA | 1016 | 7% | 0% | 46 |
| 8 | CELLULAR & MOLECULAR BIOLOGY RESEARCH | 961 | 0% | 1% | 2 |
| 9 | TRENDS IN MICROBIOLOGY | 953 | 1% | 0% | 6 |
| 10 | BMC MICROBIOLOGY | 756 | 1% | 0% | 7 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CAULOBACTER | 1455435 | 8% | 63% | 48 | Search CAULOBACTER | Search CAULOBACTER |
| 2 | CTRA | 660707 | 3% | 81% | 17 | Search CTRA | Search CTRA |
| 3 | CAULOBACTER CRESCENTUS | 550307 | 5% | 35% | 33 | Search CAULOBACTER+CRESCENTUS | Search CAULOBACTER+CRESCENTUS |
| 4 | CCRM | 216043 | 1% | 75% | 6 | Search CCRM | Search CCRM |
| 5 | GCRA | 100016 | 1% | 42% | 5 | Search GCRA | Search GCRA |
| 6 | CTRA RESPONSE REGULATOR | 96021 | 0% | 100% | 2 | Search CTRA+RESPONSE+REGULATOR | Search CTRA+RESPONSE+REGULATOR |
| 7 | PHICBK | 96021 | 0% | 100% | 2 | Search PHICBK | Search PHICBK |
| 8 | CLPXP | 69180 | 1% | 21% | 7 | Search CLPXP | Search CLPXP |
| 9 | 45 S RNA | 64012 | 0% | 67% | 2 | Search 45+S+RNA | Search 45+S+RNA |
| 10 | GENE TRANSFER AGENT | 61724 | 0% | 43% | 3 | Search GENE+TRANSFER+AGENT | Search GENE+TRANSFER+AGENT |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | CURTIS, PD , BRUN, YV , (2010) GETTING IN THE LOOP: REGULATION OF DEVELOPMENT IN CAULOBACTER CRESCENTUS.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 74. ISSUE 1. P. 13-41 | 159 | 61% | 91 |
| 2 | BROWN, PJB , HARDY, GG , TRIMBLE, MJ , BRUN, YV , (2009) COMPLEX REGULATORY PATHWAYS COORDINATE CELL-CYCLE PROGRESSION AND DEVELOPMENT IN CAULOBACTER CRESCENTUS.ADVANCES IN MICROBIAL PHYSIOLOGY, VOL 54. VOL. 54. ISSUE . P. 1-101 | 135 | 50% | 27 |
| 3 | PANIS, G , MURRAY, SR , VIOLLIER, PH , (2015) VERSATILITY OF GLOBAL TRANSCRIPTIONAL REGULATORS IN ALPHA-PROTEOBACTERIA: FROM ESSENTIAL CELL CYCLE CONTROL TO ANCILLARY FUNCTIONS.FEMS MICROBIOLOGY REVIEWS. VOL. 39. ISSUE 1. P. 120 -133 | 62 | 65% | 9 |
| 4 | TSOKOS, CG , LAUB, MT , (2012) POLARITY AND CELL FATE ASYMMETRY IN CAULOBACTER CRESCENTUS.CURRENT OPINION IN MICROBIOLOGY. VOL. 15. ISSUE 6. P. 744-750 | 47 | 89% | 38 |
| 5 | CURTIS, PD , BRUN, YV , (2014) IDENTIFICATION OF ESSENTIAL ALPHAPROTEOBACTERIAL GENES REVEALS OPERATIONAL VARIABILITY IN CONSERVED DEVELOPMENTAL AND CELL CYCLE SYSTEMS.MOLECULAR MICROBIOLOGY. VOL. 93. ISSUE 4. P. 713 -735 | 64 | 66% | 9 |
| 6 | COLLIER, J , (2012) REGULATION OF CHROMOSOMAL REPLICATION IN CAULOBACTER CRESCENTUS.PLASMID. VOL. 67. ISSUE 2. P. 76-87 | 60 | 71% | 21 |
| 7 | SANSELICIO, S , BERGE, M , THERAULAZ, L , RADHAKRISHNAN, SK , VIOLLIER, PH , (2015) TOPOLOGICAL CONTROL OF THE CAULOBACTER CELL CYCLE CIRCUITRY BY A POLARIZED SINGLE-DOMAIN PAS PROTEIN.NATURE COMMUNICATIONS. VOL. 6. ISSUE . P. - | 52 | 76% | 4 |
| 8 | JENAL, U , (2000) SIGNAL TRANSDUCTION MECHANISMS IN CAULOBACTER CRESCENTUS DEVELOPMENT AND CELL CYCLE CONTROL.FEMS MICROBIOLOGY REVIEWS. VOL. 24. ISSUE 2. P. 177 -191 | 75 | 77% | 32 |
| 9 | AUSMEES, N , JACOBS-WAGNER, C , (2003) SPATIAL AND TEMPORAL CONTROL OF DIFFERENTIATION AND CELL CYCLE PROGRESSION IN CAULOBACTER CRESCENTUS.ANNUAL REVIEW OF MICROBIOLOGY. VOL. 57. ISSUE . P. 225-247 | 59 | 79% | 57 |
| 10 | MURRAY, SM , PANIS, G , FUMEAUX, C , VIOLLIER, PH , HOWARD, M , (2013) COMPUTATIONAL AND GENETIC REDUCTION OF A CELL CYCLE TO ITS SIMPLEST, PRIMORDIAL COMPONENTS.PLOS BIOLOGY. VOL. 11. ISSUE 12. P. - | 44 | 77% | 13 |
Classes with closest relation at Level 1 |