Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
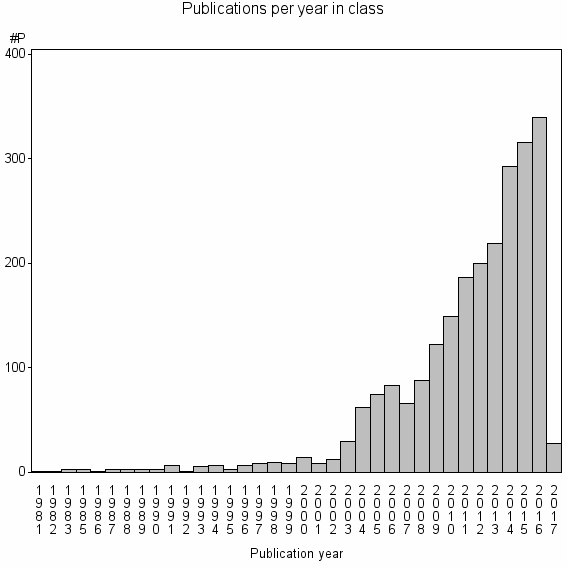
| 2108 | 2354 | 42.7 | 86% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 5 | 4 | CHEMISTRY, ORGANIC//CHEMISTRY, INORGANIC & NUCLEAR//CHEMISTRY, MULTIDISCIPLINARY | 1745167 |
| 183 | 3 | APTAMER//G QUADRUPLEX//RIBOZYME | 56307 |
| 1995 | 2 | DNA ORIGAMI//DNA NANOTECHNOLOGY//DNA NANOSTRUCTURES | 5542 |
| 2108 | 1 | DNA ORIGAMI//DNA NANOTECHNOLOGY//DNA NANOSTRUCTURES | 2354 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | DNA ORIGAMI | authKW | 1602804 | 7% | 78% | 158 |
| 2 | DNA NANOTECHNOLOGY | authKW | 1423957 | 7% | 71% | 154 |
| 3 | DNA NANOSTRUCTURES | authKW | 612822 | 3% | 71% | 67 |
| 4 | DNA TILES | authKW | 338943 | 1% | 93% | 28 |
| 5 | TILE ASSEMBLY MODEL | authKW | 304995 | 1% | 87% | 27 |
| 6 | DNA SELF ASSEMBLY | authKW | 272886 | 2% | 48% | 44 |
| 7 | MOLECULAR PROGRAMMING | authKW | 270132 | 1% | 77% | 27 |
| 8 | DNA STRUCTURES | authKW | 235024 | 3% | 23% | 80 |
| 9 | DNA STRAND DISPLACEMENT | authKW | 207897 | 1% | 70% | 23 |
| 10 | RNA NANOTECHNOLOGY | authKW | 178883 | 1% | 69% | 20 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Chemistry, Multidisciplinary | 17239 | 55% | 0% | 1287 |
| 2 | Nanoscience & Nanotechnology | 14933 | 27% | 0% | 641 |
| 3 | Materials Science, Multidisciplinary | 3412 | 29% | 0% | 672 |
| 4 | Chemistry, Physical | 1677 | 20% | 0% | 474 |
| 5 | Physics, Applied | 1229 | 17% | 0% | 405 |
| 6 | Physics, Condensed Matter | 1029 | 13% | 0% | 303 |
| 7 | Computer Science, Theory & Methods | 808 | 6% | 0% | 135 |
| 8 | Biochemistry & Molecular Biology | 242 | 13% | 0% | 307 |
| 9 | Mathematical & Computational Biology | 241 | 2% | 0% | 51 |
| 10 | Biochemical Research Methods | 211 | 4% | 0% | 97 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA NANOTECHNOL | 132812 | 1% | 39% | 26 |
| 2 | SUNGKYUNKWAN ADV NANOTECHNOL SAINT | 130734 | 1% | 31% | 33 |
| 3 | ZNN WSI | 81191 | 1% | 52% | 12 |
| 4 | BIODESIGN | 79376 | 5% | 6% | 108 |
| 5 | MOL DESIGN BIOMIMICRY | 77819 | 0% | 100% | 6 |
| 6 | DNA NANOTECHNOL CDNA | 70583 | 1% | 26% | 21 |
| 7 | LEHRSTUHL BIOELEKT | 70030 | 0% | 60% | 9 |
| 8 | CAS INTER IAL PHYS TECHNOL | 60776 | 1% | 31% | 15 |
| 9 | MOL DESIGN BIOMIMET | 55331 | 0% | 53% | 8 |
| 10 | BIOL INTER ES IBG 1 | 47542 | 0% | 33% | 11 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NATURAL COMPUTING | 51812 | 2% | 11% | 38 |
| 2 | NATURE NANOTECHNOLOGY | 21864 | 2% | 4% | 47 |
| 3 | NANO LETTERS | 18126 | 6% | 1% | 131 |
| 4 | SMALL | 14720 | 3% | 2% | 76 |
| 5 | ACS NANO | 13317 | 4% | 1% | 98 |
| 6 | ANGEWANDTE CHEMIE-INTERNATIONAL EDITION | 9388 | 7% | 0% | 169 |
| 7 | MATCH-COMMUNICATIONS IN MATHEMATICAL AND IN COMPUTER CHEMISTRY | 5845 | 1% | 2% | 26 |
| 8 | NATURE CHEMISTRY | 5645 | 1% | 2% | 21 |
| 9 | JOURNAL OF THE AMERICAN CHEMICAL SOCIETY | 5337 | 8% | 0% | 199 |
| 10 | NANOSCALE | 4557 | 3% | 1% | 60 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA ORIGAMI | 1602804 | 7% | 78% | 158 | Search DNA+ORIGAMI | Search DNA+ORIGAMI |
| 2 | DNA NANOTECHNOLOGY | 1423957 | 7% | 71% | 154 | Search DNA+NANOTECHNOLOGY | Search DNA+NANOTECHNOLOGY |
| 3 | DNA NANOSTRUCTURES | 612822 | 3% | 71% | 67 | Search DNA+NANOSTRUCTURES | Search DNA+NANOSTRUCTURES |
| 4 | DNA TILES | 338943 | 1% | 93% | 28 | Search DNA+TILES | Search DNA+TILES |
| 5 | TILE ASSEMBLY MODEL | 304995 | 1% | 87% | 27 | Search TILE+ASSEMBLY+MODEL | Search TILE+ASSEMBLY+MODEL |
| 6 | DNA SELF ASSEMBLY | 272886 | 2% | 48% | 44 | Search DNA+SELF+ASSEMBLY | Search DNA+SELF+ASSEMBLY |
| 7 | MOLECULAR PROGRAMMING | 270132 | 1% | 77% | 27 | Search MOLECULAR+PROGRAMMING | Search MOLECULAR+PROGRAMMING |
| 8 | DNA STRUCTURES | 235024 | 3% | 23% | 80 | Search DNA+STRUCTURES | Search DNA+STRUCTURES |
| 9 | DNA STRAND DISPLACEMENT | 207897 | 1% | 70% | 23 | Search DNA+STRAND+DISPLACEMENT | Search DNA+STRAND+DISPLACEMENT |
| 10 | RNA NANOTECHNOLOGY | 178883 | 1% | 69% | 20 | Search RNA+NANOTECHNOLOGY | Search RNA+NANOTECHNOLOGY |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SUN, LF , YU, L , SHEN, WQ , (2014) DNA NANOTECHNOLOGY AND ITS APPLICATIONS IN BIOMEDICAL RESEARCH.JOURNAL OF BIOMEDICAL NANOTECHNOLOGY. VOL. 10. ISSUE 9. P. 2350 -2370 | 198 | 93% | 5 |
| 2 | PINHEIRO, AV , HAN, DR , SHIH, WM , YAN, H , (2011) CHALLENGES AND OPPORTUNITIES FOR STRUCTURAL DNA NANOTECHNOLOGY.NATURE NANOTECHNOLOGY. VOL. 6. ISSUE 12. P. 763 -772 | 111 | 82% | 396 |
| 3 | ZHANG, F , NANGREAVE, J , LIU, Y , YAN, H , (2014) STRUCTURAL DNA NANOTECHNOLOGY: STATE OF THE ART AND FUTURE PERSPECTIVE.JOURNAL OF THE AMERICAN CHEMICAL SOCIETY. VOL. 136. ISSUE 32. P. 11198 -11211 | 117 | 80% | 99 |
| 4 | TINTORE, M , ERITJA, R , FABREGA, C , (2014) DNA NANOARCHITECTURES: STEPS TOWARDS BIOLOGICAL APPLICATIONS.CHEMBIOCHEM. VOL. 15. ISSUE 10. P. 1374 -1390 | 166 | 85% | 12 |
| 5 | ZHANG, Z , FU, YM , LI, BJ , FENG, GY , LI, C , FAN, CH , HE, L , (2011) SELF-ASSEMBLY-BASED STRUCTURAL DNA NANOTECHNOLOGY.CURRENT ORGANIC CHEMISTRY. VOL. 15. ISSUE 4. P. 534 -547 | 143 | 95% | 2 |
| 6 | LI, HY , LABEAN, TH , LEONG, KW , (2011) NUCLEIC ACID-BASED NANOENGINEERING: NOVEL STRUCTURES FOR BIOMEDICAL APPLICATIONS.INTERFACE FOCUS. VOL. 1. ISSUE 5. P. 702 -724 | 162 | 70% | 17 |
| 7 | TORRING, T , VOIGT, NV , NANGREAVE, J , YAN, H , GOTHELF, KV , (2011) DNA ORIGAMI: A QUANTUM LEAP FOR SELF-ASSEMBLY OF COMPLEX STRUCTURES.CHEMICAL SOCIETY REVIEWS. VOL. 40. ISSUE 12. P. 5636 -5646 | 73 | 97% | 178 |
| 8 | CHANDRASEKARAN, AR , LEVCHENKO, O , (2016) DNA NANOCAGES.CHEMISTRY OF MATERIALS. VOL. 28. ISSUE 16. P. 5569 -5581 | 104 | 92% | 0 |
| 9 | WANG, F , LU, CH , WILLNER, I , (2014) FROM CASCADED CATALYTIC NUCLEIC ACIDS TO ENZYME-DNA NANOSTRUCTURES: CONTROLLING REACTIVITY, SENSING, LOGIC OPERATIONS, AND ASSEMBLY OF COMPLEX STRUCTURES.CHEMICAL REVIEWS. VOL. 114. ISSUE 5. P. 2881 -2941 | 208 | 24% | 153 |
| 10 | CHANDRASEKARAN, AR , ANDERSON, N , KIZER, M , HALVORSEN, K , WANG, X , (2016) BEYOND THE FOLD: EMERGING BIOLOGICAL APPLICATIONS OF DNA ORIGAMI.CHEMBIOCHEM. VOL. 17. ISSUE 12. P. 1081 -1089 | 102 | 86% | 1 |
Classes with closest relation at Level 1 |