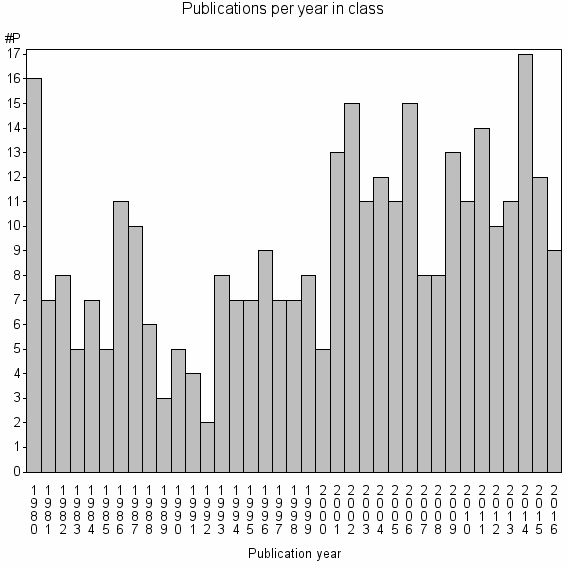
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 23719 | 337 | 32.4 | 64% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 278 | 3 | CORYNEBACTERIUM GLUTAMICUM//ACTA CRYSTALLOGRAPHICA SECTION D-BIOLOGICAL CRYSTALLOGRAPHY//BIOCHEMISTRY & MOLECULAR BIOLOGY | 43163 |
| 2061 | 2 | GLYCERALDEHYDE 3 PHOSPHATE DEHYDROGENASE//PHOSPHOFRUCTOKINASE//PHOSPHOGLYCERATE MUTASE | 5293 |
| 23719 | 1 | UDP SUGAR HYDROLASE//SUGAR PHOSPHATASES//HAD SUPERFAMILY | 337 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | UDP SUGAR HYDROLASE | authKW | 465986 | 2% | 86% | 6 |
| 2 | SUGAR PHOSPHATASES | authKW | 362435 | 1% | 100% | 4 |
| 3 | HAD SUPERFAMILY | authKW | 305199 | 2% | 42% | 8 |
| 4 | CLASS B PHOSPHATASE | authKW | 271826 | 1% | 100% | 3 |
| 5 | CLASS C ACID PHOSPHATASE | authKW | 271826 | 1% | 100% | 3 |
| 6 | MAB 1D7 | authKW | 271826 | 1% | 100% | 3 |
| 7 | BACTERIAL PHOSPHATASE | authKW | 203868 | 1% | 75% | 3 |
| 8 | PHOSPHOSERINE PHOSPHATASE | authKW | 201801 | 2% | 32% | 7 |
| 9 | ACID PHOSPHOHYDROLASES | authKW | 181218 | 1% | 100% | 2 |
| 10 | DDDD PHOSPHOHYDROLASE SUPERFAMILY | authKW | 181218 | 1% | 100% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 1300 | 52% | 0% | 174 |
| 2 | Microbiology | 938 | 23% | 0% | 77 |
| 3 | Biophysics | 423 | 14% | 0% | 46 |
| 4 | Biochemical Research Methods | 213 | 9% | 0% | 31 |
| 5 | Biotechnology & Applied Microbiology | 155 | 10% | 0% | 35 |
| 6 | Crystallography | 117 | 6% | 0% | 19 |
| 7 | Medicine, Legal | 34 | 1% | 0% | 4 |
| 8 | Food Science & Technology | 8 | 3% | 0% | 10 |
| 9 | Toxicology | 3 | 2% | 0% | 6 |
| 10 | Genetics & Heredity | 2 | 3% | 0% | 9 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOL MOL CELULAR PLANTAS EDUARDO PRIMO YUF | 120810 | 1% | 67% | 2 |
| 2 | DIPARTIMENTO BIOL MOLEC SEZ MICROBIOL | 120810 | 1% | 67% | 2 |
| 3 | ASIAN LIFE SCI | 90609 | 0% | 100% | 1 |
| 4 | BIOCHEM ROLLINS | 90609 | 0% | 100% | 1 |
| 5 | FERMENTAT BIOTECHN | 90609 | 0% | 100% | 1 |
| 6 | FUJIAN CHEM BIOL | 90609 | 0% | 100% | 1 |
| 7 | GENE FUCT DEV | 90609 | 0% | 100% | 1 |
| 8 | MED POS EDUC | 90609 | 0% | 100% | 1 |
| 9 | POLICLIN MARIA SCOTTE | 90609 | 0% | 100% | 1 |
| 10 | SYNCHROTRON SCI GRP | 90609 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JAPANESE JOURNAL OF TOXICOLOGY AND ENVIRONMENTAL HEALTH | 4380 | 1% | 1% | 5 |
| 2 | ACTA PATHOLOGICA ET MICROBIOLOGICA SCANDINAVICA SECTION B-MICROBIOLOGY | 2873 | 1% | 2% | 2 |
| 3 | ZEITSCHRIFT FUR RECHTSMEDIZIN-JOURNAL OF LEGAL MEDICINE | 2195 | 1% | 1% | 4 |
| 4 | ACTA CRYSTALLOGRAPHICA SECTION F-STRUCTURAL BIOLOGY COMMUNICATIONS | 1888 | 2% | 0% | 8 |
| 5 | ACTA CRYSTALLOGRAPHICA SECTION D-STRUCTURAL BIOLOGY | 1152 | 1% | 0% | 4 |
| 6 | BIOCHEMISTRY | 872 | 7% | 0% | 23 |
| 7 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 685 | 2% | 0% | 7 |
| 8 | ANTONIE VAN LEEUWENHOEK JOURNAL OF MICROBIOLOGY | 632 | 1% | 0% | 2 |
| 9 | BIOLOGICAL CHEMISTRY HOPPE-SEYLER | 530 | 1% | 0% | 3 |
| 10 | JOURNAL OF FERMENTATION AND BIOENGINEERING | 364 | 1% | 0% | 3 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | UDP SUGAR HYDROLASE | 465986 | 2% | 86% | 6 | Search UDP+SUGAR+HYDROLASE | Search UDP+SUGAR+HYDROLASE |
| 2 | SUGAR PHOSPHATASES | 362435 | 1% | 100% | 4 | Search SUGAR+PHOSPHATASES | Search SUGAR+PHOSPHATASES |
| 3 | HAD SUPERFAMILY | 305199 | 2% | 42% | 8 | Search HAD+SUPERFAMILY | Search HAD+SUPERFAMILY |
| 4 | CLASS B PHOSPHATASE | 271826 | 1% | 100% | 3 | Search CLASS+B+PHOSPHATASE | Search CLASS+B+PHOSPHATASE |
| 5 | CLASS C ACID PHOSPHATASE | 271826 | 1% | 100% | 3 | Search CLASS+C+ACID+PHOSPHATASE | Search CLASS+C+ACID+PHOSPHATASE |
| 6 | MAB 1D7 | 271826 | 1% | 100% | 3 | Search MAB+1D7 | Search MAB+1D7 |
| 7 | BACTERIAL PHOSPHATASE | 203868 | 1% | 75% | 3 | Search BACTERIAL+PHOSPHATASE | Search BACTERIAL+PHOSPHATASE |
| 8 | PHOSPHOSERINE PHOSPHATASE | 201801 | 2% | 32% | 7 | Search PHOSPHOSERINE+PHOSPHATASE | Search PHOSPHOSERINE+PHOSPHATASE |
| 9 | ACID PHOSPHOHYDROLASES | 181218 | 1% | 100% | 2 | Search ACID+PHOSPHOHYDROLASES | Search ACID+PHOSPHOHYDROLASES |
| 10 | DDDD PHOSPHOHYDROLASE SUPERFAMILY | 181218 | 1% | 100% | 2 | Search DDDD+PHOSPHOHYDROLASE+SUPERFAMILY | Search DDDD+PHOSPHOHYDROLASE+SUPERFAMILY |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | DUNAWAY-MARIANO, D , ALLEN, KN , (2009) MARKERS OF FITNESS IN A SUCCESSFUL ENZYME SUPERFAMILY.CURRENT OPINION IN STRUCTURAL BIOLOGY. VOL. 19. ISSUE 6. P. 658 -665 | 19 | 66% | 35 |
| 2 | HUANG, H , PATSKOVSKY, Y , TORO, R , FARELLI, JD , PANDYA, C , ALMO, SC , ALLEN, KN , DUNAWAY-MARIANO, D , (2011) DIVERGENCE OF STRUCTURE AND FUNCTION IN THE HALOACID DEHALOGENASE ENZYME SUPERFAMILY: BACTEROIDES THETAIOTAOMICRON BT2127 IS AN INORGANIC PYROPHOSPHATASE.BIOCHEMISTRY. VOL. 50. ISSUE 41. P. 8937 -8949 | 19 | 56% | 11 |
| 3 | PARK, J , GUGGISBERG, AM , ODOM, AR , TOLIA, NH , (2015) CAP-DOMAIN CLOSURE ENABLES DIVERSE SUBSTRATE RECOGNITION BY THE C2-TYPE HALOACID DEHALOGENASE-LIKE SUGAR PHOSPHATASE PLASMODIUM FALCIPARUM HAD1.ACTA CRYSTALLOGRAPHICA SECTION D-STRUCTURAL BIOLOGY. VOL. 71. ISSUE . P. 1824 -1834 | 20 | 49% | 1 |
| 4 | CALDERONE, V , FORLEO, C , BENVENUTI, M , THALLER, MC , ROSSOLINI, GM , MANGANI, S , (2006) A STRUCTURE-BASED PROPOSAL FOR THE CATALYTIC MECHANISM OF THE BACTERIAL ACID PHOSPHATASE APHA BELONGING TO THE DDDD SUPERFAMILY OF PHOSPHOHYDROLASES.JOURNAL OF MOLECULAR BIOLOGY. VOL. 355. ISSUE 4. P. 708-721 | 20 | 57% | 12 |
| 5 | CAPARROS-MARTIN, JA , MCCARTHY-SUAREZ, I , CULIANEZ-MACIA, FA , (2013) HAD HYDROLASE FUNCTION UNVEILED BY SUBSTRATE SCREENING: ENZYMATIC CHARACTERIZATION OF ARABIDOPSIS THALIANA SUBCLASS I PHOSPHOSUGAR PHOSPHATASE ATSGPP.PLANTA. VOL. 237. ISSUE 4. P. 943-954 | 18 | 50% | 8 |
| 6 | NGUYEN, HH , WANG, LB , HUANG, H , PEISACH, E , DUNAWAY-MARIANO, D , ALLEN, KN , (2010) STRUCTURAL DETERMINANTS OF SUBSTRATE RECOGNITION IN THE HAD SUPERFAMILY MEMBER D-GLYCERO-D-MANNO-HEPTOSE-1,7-BISPHOSPHATE PHOSPHATASE (GMHB).BIOCHEMISTRY. VOL. 49. ISSUE 6. P. 1082 -1092 | 19 | 51% | 11 |
| 7 | CAPARROS-MARTIN, JA , MCCARTHY-SUAREZ, I , CULIANEZ-MACIA, FA , (2014) THE KINETIC ANALYSIS OF THE SUBSTRATE SPECIFICITY OF MOTIF 5 IN A HAD HYDROLASE-TYPE PHOSPHOSUGAR PHOSPHATASE OF ARABIDOPSIS THALIANA.PLANTA. VOL. 240. ISSUE 3. P. 479 -487 | 17 | 52% | 1 |
| 8 | LAHIRI, SD , ZHANG, GF , DUNAWAY-MARIANO, D , ALLEN, KN , (2006) DIVERSIFICATION OF FUNCTION IN THE HALOACID DEHALOGENASE ENZYME SUPERFAMILY: THE ROLE OF THE CAP DOMAIN IN HYDROLYTIC PHOSPHORUS-CARBON BOND CLEAVAGE.BIOORGANIC CHEMISTRY. VOL. 34. ISSUE 6. P. 394-409 | 20 | 51% | 16 |
| 9 | SEIFRIED, A , BERGERON, A , BOIVIN, B , GOHLA, A , (2016) REVERSIBLE OXIDATION CONTROLS THE ACTIVITY AND OLIGOMERIC STATE OF THE MAMMALIAN PHOSPHOGLYCOLATE PHOSPHATASE AUM.FREE RADICAL BIOLOGY AND MEDICINE. VOL. 97. ISSUE . P. 75 -84 | 18 | 39% | 2 |
| 10 | SINGH, H , MALINSKI, TJ , REILLY, TJ , HENZL, MT , TANNER, JJ , (2011) CRYSTAL STRUCTURE AND IMMUNOGENICITY OF THE CLASS C ACID PHOSPHATASE FROM PASTEURELLA MULTOCIDA.ARCHIVES OF BIOCHEMISTRY AND BIOPHYSICS. VOL. 509. ISSUE 1. P. 76 -81 | 15 | 52% | 2 |
Classes with closest relation at Level 1 |