Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
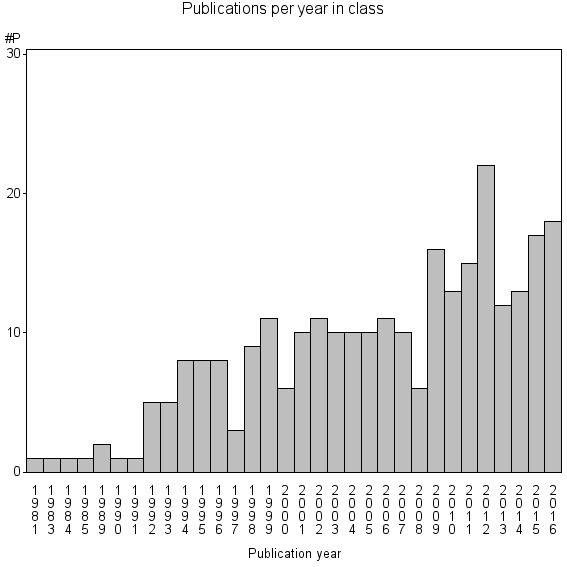
| 25675 | 275 | 38.5 | 81% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 1067 | 2 | ANGIOGENIN//RIBONUCLEASE//MOLTEN GLOBULE | 9981 |
| 25675 | 1 | PROTEIN SWITCH//CIRCULAR PERMUTATION//BIOMED ST PAU IIB SANTPAU | 275 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN SWITCH | authKW | 639563 | 4% | 48% | 12 |
| 2 | CIRCULAR PERMUTATION | authKW | 543841 | 8% | 21% | 23 |
| 3 | BIOMED ST PAU IIB SANTPAU | address | 355318 | 1% | 80% | 4 |
| 4 | ONCOGENESIS ANTITUMOR DRUG GRP | address | 307482 | 2% | 46% | 6 |
| 5 | DOMAIN INSERTION | authKW | 231322 | 2% | 42% | 5 |
| 6 | ALTERNATE FRAME FOLDING | authKW | 222075 | 1% | 100% | 2 |
| 7 | BIOMED ST PAU IIB STPAU | address | 222075 | 1% | 100% | 2 |
| 8 | PERMUTEIN | authKW | 222075 | 1% | 100% | 2 |
| 9 | REDUNDANT AND DELETED RESIDUES | authKW | 222075 | 1% | 100% | 2 |
| 10 | ARTIFICIAL VIRUSES | authKW | 222071 | 1% | 50% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 995 | 50% | 0% | 138 |
| 2 | Biotechnology & Applied Microbiology | 377 | 17% | 0% | 46 |
| 3 | Biophysics | 194 | 11% | 0% | 29 |
| 4 | Biochemical Research Methods | 101 | 7% | 0% | 20 |
| 5 | Chemistry, Multidisciplinary | 81 | 14% | 0% | 38 |
| 6 | Water Resources | 76 | 5% | 0% | 14 |
| 7 | Nanoscience & Nanotechnology | 49 | 5% | 0% | 15 |
| 8 | Chemistry, Medicinal | 19 | 3% | 0% | 9 |
| 9 | Cell Biology | 9 | 5% | 0% | 14 |
| 10 | Medicine, Research & Experimental | 8 | 3% | 0% | 9 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOMED ST PAU IIB SANTPAU | 355318 | 1% | 80% | 4 |
| 2 | ONCOGENESIS ANTITUMOR DRUG GRP | 307482 | 2% | 46% | 6 |
| 3 | BIOMED ST PAU IIB STPAU | 222075 | 1% | 100% | 2 |
| 4 | BIOTECNOL BIOMED | 210167 | 13% | 5% | 35 |
| 5 | INTER PROGRAM BIOMOL SCI ENGN | 144187 | 4% | 13% | 10 |
| 6 | BIOL FONAMENTAL | 137818 | 4% | 10% | 12 |
| 7 | BIOSENSORS NANOMACHINES | 118434 | 1% | 27% | 4 |
| 8 | AUSTRALIAN BIOENGN NANOTECLMOL | 111037 | 0% | 100% | 1 |
| 9 | BIOCHEM FUNDAMENTAL PL MOL EVOLUT | 111037 | 0% | 100% | 1 |
| 10 | BIOCHIM GENET MOL BACTERIENNE | 111037 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROTEIN ENGINEERING DESIGN & SELECTION | 3839 | 2% | 1% | 6 |
| 2 | PROTEIN SCIENCE | 3455 | 5% | 0% | 14 |
| 3 | ACS SYNTHETIC BIOLOGY | 1788 | 1% | 1% | 3 |
| 4 | GROUND WATER | 1408 | 2% | 0% | 6 |
| 5 | NANOMEDICINE-NANOTECHNOLOGY BIOLOGY AND MEDICINE | 1379 | 1% | 0% | 4 |
| 6 | PROTEIN ENGINEERING | 1220 | 2% | 0% | 5 |
| 7 | BIOTECHNOLOGY AND BIOENGINEERING | 756 | 3% | 0% | 9 |
| 8 | TRENDS IN BIOTECHNOLOGY | 720 | 1% | 0% | 4 |
| 9 | CHEMBIOCHEM | 616 | 2% | 0% | 5 |
| 10 | MOLECULAR THERAPY-METHODS & CLINICAL DEVELOPMENT | 608 | 0% | 1% | 1 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN SWITCH | 639563 | 4% | 48% | 12 | Search PROTEIN+SWITCH | Search PROTEIN+SWITCH |
| 2 | CIRCULAR PERMUTATION | 543841 | 8% | 21% | 23 | Search CIRCULAR+PERMUTATION | Search CIRCULAR+PERMUTATION |
| 3 | DOMAIN INSERTION | 231322 | 2% | 42% | 5 | Search DOMAIN+INSERTION | Search DOMAIN+INSERTION |
| 4 | ALTERNATE FRAME FOLDING | 222075 | 1% | 100% | 2 | Search ALTERNATE+FRAME+FOLDING | Search ALTERNATE+FRAME+FOLDING |
| 5 | PERMUTEIN | 222075 | 1% | 100% | 2 | Search PERMUTEIN | Search PERMUTEIN |
| 6 | REDUNDANT AND DELETED RESIDUES | 222075 | 1% | 100% | 2 | Search REDUNDANT+AND+DELETED+RESIDUES | Search REDUNDANT+AND+DELETED+RESIDUES |
| 7 | ARTIFICIAL VIRUSES | 222071 | 1% | 50% | 4 | Search ARTIFICIAL+VIRUSES | Search ARTIFICIAL+VIRUSES |
| 8 | ALLOSTERIC BIOSENSOR | 111037 | 0% | 100% | 1 | Search ALLOSTERIC+BIOSENSOR | Search ALLOSTERIC+BIOSENSOR |
| 9 | ALLOSTERIC PROTEIN SWITCHES | 111037 | 0% | 100% | 1 | Search ALLOSTERIC+PROTEIN+SWITCHES | Search ALLOSTERIC+PROTEIN+SWITCHES |
| 10 | AMPEROMETRY OF 4 AMINOPHENOL | 111037 | 0% | 100% | 1 | Search AMPEROMETRY+OF+4+AMINOPHENOL | Search AMPEROMETRY+OF+4+AMINOPHENOL |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | STRATTON, MM , LOH, SN , (2011) CONVERTING A PROTEIN INTO A SWITCH FOR BIOSENSING AND FUNCTIONAL REGULATION.PROTEIN SCIENCE. VOL. 20. ISSUE 1. P. 19 -29 | 28 | 35% | 24 |
| 2 | YU, Y , LUTZ, S , (2011) CIRCULAR PERMUTATION: A DIFFERENT WAY TO ENGINEER ENZYME STRUCTURE AND FUNCTION.TRENDS IN BIOTECHNOLOGY. VOL. 29. ISSUE 1. P. 18-25 | 26 | 34% | 40 |
| 3 | FERRAZ, RM , VERA, A , ARIS, A , VILLAVERDE, A , (2006) INSERTIONAL PROTEIN ENGINEERING FOR ANALYTICAL MOLECULAR SENSING.MICROBIAL CELL FACTORIES. VOL. 5. ISSUE . P. - | 25 | 44% | 11 |
| 4 | BANALA, S , ARTS, R , APER, SJA , MERKX, M , (2013) NO WASHING, LESS WAITING: ENGINEERING BIOMOLECULAR REPORTERS FOR SINGLE-STEP ANTIBODY DETECTION IN SOLUTION.ORGANIC & BIOMOLECULAR CHEMISTRY. VOL. 11. ISSUE 44. P. 7642-7649 | 18 | 47% | 4 |
| 5 | KANWAR, M , WRIGHT, RC , DATE, A , TULLMAN, J , OSTERMEIER, M , (2013) PROTEIN SWITCH ENGINEERING BY DOMAIN INSERTION.METHODS IN PROTEIN DESIGN. VOL. 523. ISSUE . P. 369-388 | 12 | 67% | 10 |
| 6 | CAZORLA, D , FELIU, JX , FERRER-MIRALLES, N , VILLAVERDE, A , (2002) TAILORING MOLECULAR SENSING FOR PEPTIDE DISPLAYING ENGINEERED ENZYMES.BIOTECHNOLOGY LETTERS. VOL. 24. ISSUE 6. P. 469-477 | 16 | 70% | 1 |
| 7 | WANG, Q , WANG, Y , CHEN, GJ , (2016) INFLUENCE OF SECONDARY-STRUCTURE FOLDING ON THE MUTUALLY EXCLUSIVE FOLDING PROCESS OF GL5/I27 PROTEIN: EVIDENCE FROM MOLECULAR DYNAMICS SIMULATIONS.INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. VOL. 17. ISSUE 11. P. - | 21 | 37% | 0 |
| 8 | GOLYNSKIY, MV , KOAY, MS , VINKENBORG, JL , MERKX, M , (2011) ENGINEERING PROTEIN SWITCHES: SENSORS, REGULATORS, AND SPARE PARTS FOR BIOLOGY AND BIOTECHNOLOGY.CHEMBIOCHEM. VOL. 12. ISSUE 3. P. 353-361 | 28 | 28% | 23 |
| 9 | PIERRE, B , SHAH, V , XIAO, J , KIM, JR , (2015) CONSTRUCTION OF A RANDOM CIRCULAR PERMUTATION LIBRARY USING AN ENGINEERED TRANSPOSON.ANALYTICAL BIOCHEMISTRY. VOL. 474. ISSUE . P. 16 -24 | 17 | 45% | 1 |
| 10 | FERRAZ, RM , ARIS, A , VILLAVERDE, A , (2006) ENHANCED MOLECULAR RECOGNITION SIGNAL IN ALLOSTERIC BIOSENSING BY PROPER SUBSTRATE SELECTION.BIOTECHNOLOGY AND BIOENGINEERING. VOL. 94. ISSUE 1. P. 193-199 | 12 | 80% | 4 |
Classes with closest relation at Level 1 |