Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
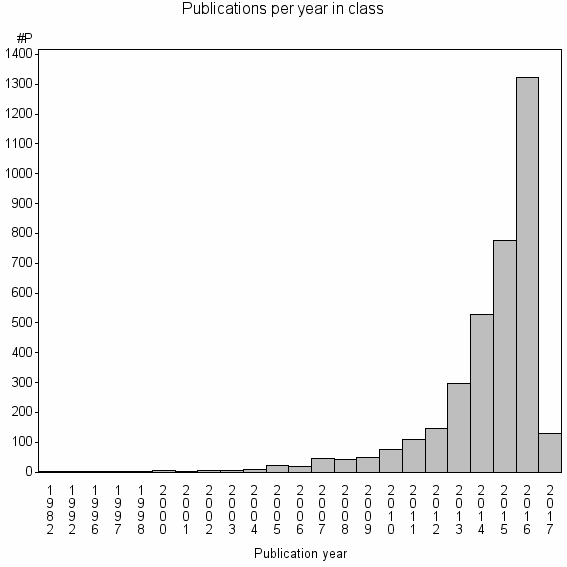
| 357 | 3591 | 52.5 | 93% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 227 | 3 | MICRORNA//MIRNA//GENE DELIVERY | 49432 |
| 57 | 2 | MICRORNA//MIRNA//LONG NONCODING RNAS | 29594 |
| 357 | 1 | LONG NONCODING RNAS//LNCRNA//LONG NONCODING RNA | 3591 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | LONG NONCODING RNAS | authKW | 3676826 | 17% | 73% | 593 |
| 2 | LNCRNA | authKW | 3074108 | 14% | 71% | 506 |
| 3 | LONG NONCODING RNA | authKW | 2027433 | 9% | 75% | 318 |
| 4 | MALAT1 | authKW | 908508 | 3% | 88% | 121 |
| 5 | HOTAIR | authKW | 888342 | 3% | 89% | 117 |
| 6 | UCA1 | authKW | 401005 | 1% | 94% | 50 |
| 7 | INCRNA | authKW | 386036 | 2% | 64% | 71 |
| 8 | LINCRNA | authKW | 313766 | 2% | 68% | 54 |
| 9 | NEAT1 | authKW | 300706 | 1% | 91% | 39 |
| 10 | GAS5 | authKW | 251982 | 1% | 87% | 34 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Oncology | 18213 | 37% | 0% | 1344 |
| 2 | Cell Biology | 9905 | 28% | 0% | 998 |
| 3 | Genetics & Heredity | 5231 | 17% | 0% | 606 |
| 4 | Biochemistry & Molecular Biology | 3849 | 30% | 0% | 1071 |
| 5 | Medicine, Research & Experimental | 1063 | 8% | 0% | 270 |
| 6 | Biotechnology & Applied Microbiology | 987 | 8% | 0% | 302 |
| 7 | Pathology | 454 | 4% | 0% | 139 |
| 8 | Developmental Biology | 189 | 2% | 0% | 66 |
| 9 | Biophysics | 171 | 4% | 0% | 131 |
| 10 | Cell & Tissue Engineering | 142 | 1% | 0% | 24 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMAT SCI TECHNOL | 61534 | 2% | 13% | 58 |
| 2 | FUNCT RNOM TEAM | 49459 | 0% | 73% | 8 |
| 3 | NINGBO YINZHOU PEOPLES HOSP | 49459 | 0% | 73% | 8 |
| 4 | PROGRAM EPITHELIAL BIOL | 47441 | 1% | 15% | 37 |
| 5 | MED REPROGRAMMING TECHNOL | 39552 | 0% | 42% | 11 |
| 6 | NEUROSCI NEUROL OPHTHALMOL | 35421 | 0% | 83% | 5 |
| 7 | ZHEJIANG PATHOPHYSIOL | 34002 | 0% | 67% | 6 |
| 8 | AFFILIATED HOSP 1 | 27890 | 12% | 1% | 416 |
| 9 | HELMHOLTZ UNIV GRP MOL RNA BIOL CANC | 25899 | 0% | 38% | 8 |
| 10 | ADV COMP COMP TECHNOL | 25504 | 0% | 100% | 3 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ONCOTARGET | 45615 | 7% | 2% | 257 |
| 2 | TUMOR BIOLOGY | 29746 | 4% | 2% | 147 |
| 3 | INTERNATIONAL JOURNAL OF CLINICAL AND EXPERIMENTAL PATHOLOGY | 14349 | 3% | 2% | 98 |
| 4 | MOLECULAR CANCER | 6881 | 1% | 2% | 38 |
| 5 | BRIEFINGS IN FUNCTIONAL GENOMICS | 5314 | 0% | 4% | 14 |
| 6 | RNA BIOLOGY | 5293 | 1% | 3% | 24 |
| 7 | AMERICAN JOURNAL OF TRANSLATIONAL RESEARCH | 5236 | 1% | 2% | 26 |
| 8 | FRONTIERS IN GENETICS | 5107 | 1% | 3% | 22 |
| 9 | ONCOTARGETS AND THERAPY | 5090 | 1% | 2% | 34 |
| 10 | CELL DEATH & DISEASE | 3193 | 1% | 1% | 30 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | LONG NONCODING RNAS | 3676826 | 17% | 73% | 593 | Search LONG+NONCODING+RNAS | Search LONG+NONCODING+RNAS |
| 2 | LNCRNA | 3074108 | 14% | 71% | 506 | Search LNCRNA | Search LNCRNA |
| 3 | LONG NONCODING RNA | 2027433 | 9% | 75% | 318 | Search LONG+NONCODING+RNA | Search LONG+NONCODING+RNA |
| 4 | MALAT1 | 908508 | 3% | 88% | 121 | Search MALAT1 | Search MALAT1 |
| 5 | HOTAIR | 888342 | 3% | 89% | 117 | Search HOTAIR | Search HOTAIR |
| 6 | UCA1 | 401005 | 1% | 94% | 50 | Search UCA1 | Search UCA1 |
| 7 | INCRNA | 386036 | 2% | 64% | 71 | Search INCRNA | Search INCRNA |
| 8 | LINCRNA | 313766 | 2% | 68% | 54 | Search LINCRNA | Search LINCRNA |
| 9 | NEAT1 | 300706 | 1% | 91% | 39 | Search NEAT1 | Search NEAT1 |
| 10 | GAS5 | 251982 | 1% | 87% | 34 | Search GAS5 | Search GAS5 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SCHMITT, AM , CHANG, HY , (2016) LONG NONCODING RNAS IN CANCER PATHWAYS.CANCER CELL. VOL. 29. ISSUE 4. P. 452 -463 | 119 | 73% | 26 |
| 2 | DHAMIJA, S , DIEDERICHS, S , (2016) FROM JUNK TO MASTER REGULATORS OF INVASION: LNCRNA FUNCTIONS IN MIGRATION, EMT AND METASTASIS.INTERNATIONAL JOURNAL OF CANCER. VOL. 139. ISSUE 2. P. 269 -280 | 137 | 87% | 9 |
| 3 | SCHMITZ, SU , GROTE, P , HERRMANN, BG , (2016) MECHANISMS OF LONG NONCODING RNA FUNCTION IN DEVELOPMENT AND DISEASE.CELLULAR AND MOLECULAR LIFE SCIENCES. VOL. 73. ISSUE 13. P. 2491 -2509 | 124 | 70% | 17 |
| 4 | ZHANG, F , ZHANG, L , ZHANG, CG , (2016) LONG NONCODING RNAS AND TUMORIGENESIS: GENETIC ASSOCIATIONS, MOLECULAR MECHANISMS, AND THERAPEUTIC STRATEGIES.TUMOR BIOLOGY. VOL. 37. ISSUE 1. P. 163 -175 | 116 | 76% | 9 |
| 5 | KLADI-SKANDALI, A , MICHAELIDOU, K , SCORILAS, A , MAVRIDIS, K , (2015) LONG NONCODING RNAS IN DIGESTIVE SYSTEM MALIGNANCIES: A NOVEL CLASS OF CANCER BIOMARKERS AND THERAPEUTIC TARGETS?.GASTROENTEROLOGY RESEARCH AND PRACTICE. VOL. . ISSUE . P. - | 131 | 90% | 7 |
| 6 | HUARTE, M , (2015) THE EMERGING ROLE OF INCRNAS IN CANCER.NATURE MEDICINE. VOL. 21. ISSUE 11. P. 1253 -1261 | 94 | 76% | 63 |
| 7 | ROTH, A , DIEDERICHS, S , (2016) LONG NONCODING RNAS IN LUNG CANCER.LONG NON-CODING RNAS IN HUMAN DISEASE. VOL. 394. ISSUE . P. 57 -110 | 176 | 58% | 3 |
| 8 | FATICA, A , BOZZONI, I , (2014) LONG NON-CODING RNAS: NEW PLAYERS IN CELL DIFFERENTIATION AND DEVELOPMENT.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 1. P. 7-21 | 74 | 60% | 367 |
| 9 | SUN, M , KRAUS, WL , (2015) FROM DISCOVERY TO FUNCTION: THE EXPANDING ROLES OF LONG NONCODING RNAS IN PHYSIOLOGY AND DISEASE.ENDOCRINE REVIEWS. VOL. 36. ISSUE 1. P. 25 -64 | 171 | 54% | 21 |
| 10 | ULITSKY, I , BARTEL, DP , (2013) LINCRNAS: GENOMICS, EVOLUTION, AND MECHANISMS.CELL. VOL. 154. ISSUE 1. P. 26 -46 | 93 | 53% | 430 |
Classes with closest relation at Level 1 |