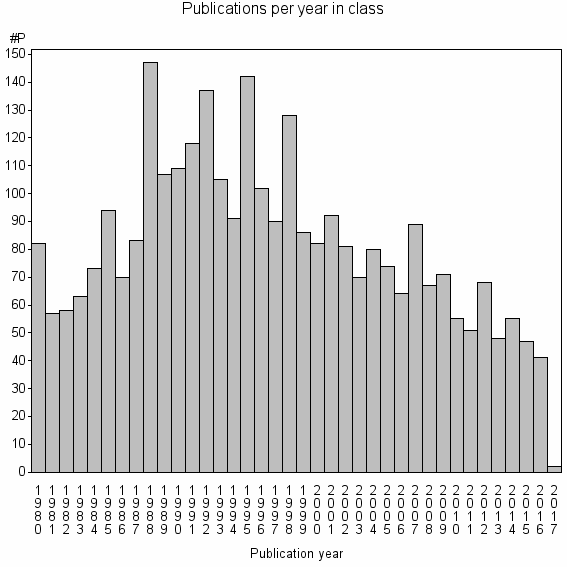
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 784 | 3079 | 35.4 | 68% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 1609 | 2 | RESTRICTION ENDONUCLEASE//BIOTECHNIQUES//RESTRICTION MODIFICATION SYSTEM | 7100 |
| 784 | 1 | RESTRICTION ENDONUCLEASE//RESTRICTION MODIFICATION SYSTEM//RESTRICTION MODIFICATION | 3079 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RESTRICTION ENDONUCLEASE | authKW | 587319 | 5% | 38% | 157 |
| 2 | RESTRICTION MODIFICATION SYSTEM | authKW | 502659 | 3% | 65% | 78 |
| 3 | RESTRICTION MODIFICATION | authKW | 452418 | 2% | 62% | 74 |
| 4 | DNA PROT INTERACT UNIT | address | 210692 | 1% | 51% | 42 |
| 5 | ISOSCHIZOMER | authKW | 191496 | 1% | 74% | 26 |
| 6 | DNA METHYLTRANSFERASE | authKW | 151603 | 3% | 14% | 106 |
| 7 | RESTRICTION ENZYME | authKW | 148374 | 2% | 26% | 57 |
| 8 | SITE SPECIFIC ENDONUCLEASE | authKW | 127826 | 1% | 68% | 19 |
| 9 | NUCLEIC ACIDS RESEARCH | journal | 111084 | 22% | 2% | 664 |
| 10 | ANTIRESTRICTION | authKW | 99979 | 0% | 92% | 11 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 25221 | 73% | 0% | 2239 |
| 2 | Genetics & Heredity | 2893 | 14% | 0% | 427 |
| 3 | Microbiology | 1632 | 11% | 0% | 335 |
| 4 | Biophysics | 1091 | 8% | 0% | 241 |
| 5 | Biochemical Research Methods | 289 | 4% | 0% | 129 |
| 6 | Cell Biology | 205 | 6% | 0% | 194 |
| 7 | Biotechnology & Applied Microbiology | 128 | 4% | 0% | 132 |
| 8 | Crystallography | 10 | 1% | 0% | 36 |
| 9 | Biology | 9 | 1% | 0% | 38 |
| 10 | Literature, Romance | 3 | 0% | 0% | 4 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA PROT INTERACT UNIT | 210692 | 1% | 51% | 42 |
| 2 | BIOINFORMAT PROT ENGN | 65600 | 1% | 19% | 35 |
| 3 | PROGRAM BIOPHYS BIOCHEM | 46194 | 1% | 27% | 17 |
| 4 | BIOPHYS S | 40638 | 1% | 23% | 18 |
| 5 | ABT BIOPHYS BIOCHEM VERFAHREN | 31728 | 0% | 80% | 4 |
| 6 | IBBS BIOPHYS S | 30982 | 0% | 63% | 5 |
| 7 | ABT STRUKTURANAL | 29746 | 0% | 100% | 3 |
| 8 | INTER PROGRAM BIOCHEM MOL BIOL | 28572 | 0% | 41% | 7 |
| 9 | BIOL DNA MODIFICAT | 24325 | 0% | 27% | 9 |
| 10 | BIOREC PROGRAM | 19831 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NUCLEIC ACIDS RESEARCH | 111084 | 22% | 2% | 664 |
| 2 | GENE | 41733 | 9% | 1% | 286 |
| 3 | JOURNAL OF MOLECULAR BIOLOGY | 15483 | 6% | 1% | 192 |
| 4 | MOLECULAR BIOLOGY | 9387 | 2% | 1% | 67 |
| 5 | BIOCHEMISTRY-MOSCOW | 7193 | 2% | 1% | 73 |
| 6 | BIOLOGICAL CHEMISTRY | 6487 | 1% | 1% | 46 |
| 7 | BIOTECHNOLOGY SOFTWARE JOURNAL | 4957 | 0% | 50% | 1 |
| 8 | BIOORGANICHESKAYA KHIMIYA | 3060 | 1% | 1% | 32 |
| 9 | BIOCHEMISTRY | 2905 | 4% | 0% | 129 |
| 10 | JOURNAL OF BACTERIOLOGY | 2835 | 3% | 0% | 100 |
Author Key Words |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | PINGOUD, A , WILSON, GG , WENDE, W , (2014) TYPE II RESTRICTION ENDONUCLEASES-A HISTORICAL PERSPECTIVE AND MORE.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE 12. P. 7489 -7527 | 306 | 82% | 26 |
| 2 | LOENEN, WAM , DRYDEN, DTF , RALEIGH, EA , WILSON, GG , (2014) TYPE I RESTRICTION ENZYMES AND THEIR RELATIVES.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE 1. P. 20 -44 | 167 | 83% | 31 |
| 3 | MCCLELLAND, M , NELSON, M , RASCHKE, E , (1994) EFFECT OF SITE-SPECIFIC MODIFICATION ON RESTRICTION ENDONUCLEASES AND DNA MODIFICATION METHYLTRANSFERASES.NUCLEIC ACIDS RESEARCH. VOL. 22. ISSUE 17. P. 3640-3659 | 268 | 83% | 276 |
| 4 | KESSLER, C , MANTA, V , (1990) SPECIFICITY OF RESTRICTION ENDONUCLEASES AND DNA MODIFICATION METHYLTRANSFERASES - A REVIEW.GENE. VOL. 92. ISSUE 1-2. P. 1-248 | 378 | 81% | 112 |
| 5 | MADHUSOODANAN, UK , RAO, DN , (2010) DIVERSITY OF DNA METHYLTRANSFERASES THAT RECOGNIZE ASYMMETRIC TARGET SEQUENCES.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 45. ISSUE 2. P. 125 -145 | 155 | 90% | 11 |
| 6 | NELSON, M , RASCHKE, E , MCCLELLAND, M , (1993) EFFECT OF SITE-SPECIFIC METHYLATION ON RESTRICTION ENDONUCLEASES AND DNA MODIFICATION METHYLTRANSFERASES.NUCLEIC ACIDS RESEARCH. VOL. 21. ISSUE 13. P. 3139-3154 | 244 | 84% | 62 |
| 7 | PINGOUD, A , FUXREITER, M , PINGOUD, V , WENDE, W , (2005) TYPE II RESTRICTION ENDONUCLEASES: STRUCTURE AND MECHANISM.CELLULAR AND MOLECULAR LIFE SCIENCES. VOL. 62. ISSUE 6. P. 685-707 | 140 | 73% | 275 |
| 8 | PINGOUD, A , JELTSCH, A , (2001) STRUCTURE AND FUNCTION OF TYPE II RESTRICTION ENDONUCLEASES.NUCLEIC ACIDS RESEARCH. VOL. 29. ISSUE 18. P. 3705-3727 | 135 | 77% | 357 |
| 9 | LOENEN, WAM , DRYDEN, DTF , RALEIGH, EA , WILSON, GG , MURRAY, NE , (2014) HIGHLIGHTS OF THE DNA CUTTERS: A SHORT HISTORY OF THE RESTRICTION ENZYMES.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE 1. P. 3 -19 | 117 | 68% | 39 |
| 10 | RAO, DN , SAHA, S , KRISHNAMURTHY, V , (2000) ATP-DEPENDENT RESTRICTION ENZYMES.PROGRESS IN NUCLEIC ACID RESEARCH AND MOLECULAR BIOLOGY, VOL 64. VOL. 64. ISSUE . P. 1 -63 | 168 | 88% | 27 |
Classes with closest relation at Level 1 |