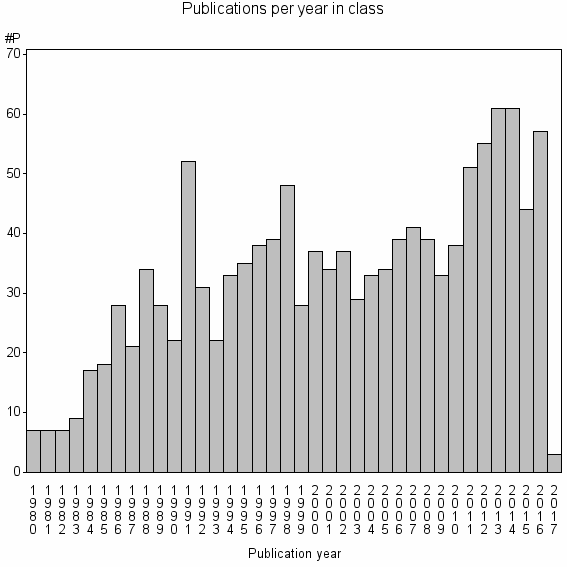
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 8466 | 1250 | 47.8 | 87% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 156 | 2 | NUCLEAR PORE COMPLEX//PRE MRNA SPLICING//SPLICEOSOME | 22915 |
| 8466 | 1 | POLYADENYLATION//ALTERNATIVE POLYADENYLATION//POLYA POLYMERASE | 1250 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | POLYADENYLATION | authKW | 922150 | 11% | 27% | 142 |
| 2 | ALTERNATIVE POLYADENYLATION | authKW | 716614 | 5% | 45% | 65 |
| 3 | POLYA POLYMERASE | authKW | 352674 | 3% | 38% | 38 |
| 4 | POLYA SITE | authKW | 323291 | 1% | 88% | 15 |
| 5 | MRNA 3 END PROCESSING | authKW | 246300 | 1% | 92% | 11 |
| 6 | 3 END PROCESSING | authKW | 246207 | 2% | 40% | 25 |
| 7 | 3 END FORMATION | authKW | 238288 | 2% | 49% | 20 |
| 8 | CLEAVAGE AND POLYADENYLATION | authKW | 219834 | 1% | 75% | 12 |
| 9 | PRE MRNA 3 END PROCESSING | authKW | 219834 | 1% | 75% | 12 |
| 10 | CLEAVAGE STIMULATION FACTOR | authKW | 146560 | 0% | 100% | 6 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 9264 | 69% | 0% | 868 |
| 2 | Cell Biology | 7809 | 41% | 0% | 508 |
| 3 | Genetics & Heredity | 1529 | 16% | 0% | 195 |
| 4 | Developmental Biology | 613 | 5% | 0% | 60 |
| 5 | Biotechnology & Applied Microbiology | 266 | 8% | 0% | 95 |
| 6 | Virology | 120 | 3% | 0% | 36 |
| 7 | Mathematical & Computational Biology | 116 | 2% | 0% | 26 |
| 8 | Biophysics | 46 | 3% | 0% | 42 |
| 9 | Biochemical Research Methods | 40 | 3% | 0% | 36 |
| 10 | Plant Sciences | 19 | 3% | 0% | 43 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MINIST EDUC COSTAL WETLAND ECOSYST | 122133 | 0% | 100% | 5 |
| 2 | P ANIKOLAOU | 79936 | 0% | 55% | 6 |
| 3 | P ANIKOLAOU ONCOL | 55829 | 0% | 57% | 4 |
| 4 | INSERM ERM 206 | 48853 | 0% | 100% | 2 |
| 5 | CELL ACTIVAT GENE EXP S GRP | 36637 | 0% | 50% | 3 |
| 6 | BIOZENTRUM CELL BIOL | 32568 | 0% | 67% | 2 |
| 7 | DIGNA BIOTECH | 32568 | 0% | 67% | 2 |
| 8 | STATE BIOCONTROLGUANGDONG PROV P | 31402 | 0% | 43% | 3 |
| 9 | BARTHOLIN BLDG | 24427 | 0% | 100% | 1 |
| 10 | BIOCHEM LIFE SCISTATE BIOCONTRO | 24427 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR AND CELLULAR BIOLOGY | 21171 | 11% | 1% | 139 |
| 2 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 13018 | 1% | 4% | 14 |
| 3 | RNA-A PUBLICATION OF THE RNA SOCIETY | 10121 | 2% | 2% | 23 |
| 4 | NUCLEIC ACIDS RESEARCH | 9877 | 10% | 0% | 127 |
| 5 | RNA | 9820 | 3% | 1% | 32 |
| 6 | GENES & DEVELOPMENT | 7506 | 4% | 1% | 48 |
| 7 | EMBO JOURNAL | 5780 | 5% | 0% | 64 |
| 8 | MOLECULAR CELL | 5436 | 3% | 1% | 35 |
| 9 | GENOME RESEARCH | 2536 | 1% | 1% | 12 |
| 10 | CELL | 2304 | 3% | 0% | 38 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | POLYADENYLATION | 922150 | 11% | 27% | 142 | Search POLYADENYLATION | Search POLYADENYLATION |
| 2 | ALTERNATIVE POLYADENYLATION | 716614 | 5% | 45% | 65 | Search ALTERNATIVE+POLYADENYLATION | Search ALTERNATIVE+POLYADENYLATION |
| 3 | POLYA POLYMERASE | 352674 | 3% | 38% | 38 | Search POLYA+POLYMERASE | Search POLYA+POLYMERASE |
| 4 | POLYA SITE | 323291 | 1% | 88% | 15 | Search POLYA+SITE | Search POLYA+SITE |
| 5 | MRNA 3 END PROCESSING | 246300 | 1% | 92% | 11 | Search MRNA+3+END+PROCESSING | Search MRNA+3+END+PROCESSING |
| 6 | 3 END PROCESSING | 246207 | 2% | 40% | 25 | Search 3+END+PROCESSING | Search 3+END+PROCESSING |
| 7 | 3 END FORMATION | 238288 | 2% | 49% | 20 | Search 3+END+FORMATION | Search 3+END+FORMATION |
| 8 | CLEAVAGE AND POLYADENYLATION | 219834 | 1% | 75% | 12 | Search CLEAVAGE+AND+POLYADENYLATION | Search CLEAVAGE+AND+POLYADENYLATION |
| 9 | PRE MRNA 3 END PROCESSING | 219834 | 1% | 75% | 12 | Search PRE+MRNA+3+END+PROCESSING | Search PRE+MRNA+3+END+PROCESSING |
| 10 | CLEAVAGE STIMULATION FACTOR | 146560 | 0% | 100% | 6 | Search CLEAVAGE+STIMULATION+FACTOR | Search CLEAVAGE+STIMULATION+FACTOR |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ZHAO, J , HYMAN, L , MOORE, C , (1999) FORMATION OF MRNA 3 ' ENDS IN EUKARYOTES: MECHANISM, REGULATION, AND INTERRELATIONSHIPS WITH OTHER STEPS IN MRNA SYNTHESIS.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 63. ISSUE 2. P. 405 -+ | 221 | 46% | 692 |
| 2 | XIANG, KH , TONG, L , MANLEY, JL , (2014) DELINEATING THE STRUCTURAL BLUEPRINT OF THE PRE-MRNA 3 '-END PROCESSING MACHINERY.MOLECULAR AND CELLULAR BIOLOGY. VOL. 34. ISSUE 11. P. 1894-1910 | 143 | 74% | 12 |
| 3 | MANDEL, CR , BAI, Y , TONG, L , (2008) PROTEIN FACTORS IN PRE-MRNA 3'-END PROCESSING.CELLULAR AND MOLECULAR LIFE SCIENCES. VOL. 65. ISSUE 7-8. P. 1099-1122 | 141 | 64% | 200 |
| 4 | TIAN, B , GRABER, JH , (2012) SIGNALS FOR PRE-MRNA CLEAVAGE AND POLYADENYLATION.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 3. ISSUE 3. P. 385 -396 | 88 | 91% | 39 |
| 5 | MILLEVOI, S , VAGNER, S , (2010) MOLECULAR MECHANISMS OF EUKARYOTIC PRE-MRNA 3' END PROCESSING REGULATION.NUCLEIC ACIDS RESEARCH. VOL. 38. ISSUE 9. P. 2757 -2774 | 111 | 62% | 160 |
| 6 | PROUDFOOT, NJ , (2011) ENDING THE MESSAGE: POLY(A) SIGNALS THEN AND NOW.GENES & DEVELOPMENT. VOL. 25. ISSUE 17. P. 1770 -1782 | 80 | 69% | 205 |
| 7 | OGORODNIKOV, A , KARGAPOLOVA, Y , DANCKWARDT, S , (2016) PROCESSING AND TRANSCRIPTOME EXPANSION AT THE MRNA 3' END IN HEALTH AND DISEASE: FINDING THE RIGHT END.PFLUGERS ARCHIV-EUROPEAN JOURNAL OF PHYSIOLOGY. VOL. 468. ISSUE 6. P. 993 -1012 | 110 | 59% | 0 |
| 8 | SHI, YS , MANLEY, JL , (2015) THE END OF THE MESSAGE: MULTIPLE PROTEIN-RNA INTERACTIONS DEFINE THE MRNA POLYADENYLATION SITE.GENES & DEVELOPMENT. VOL. 29. ISSUE 9. P. 889 -897 | 69 | 66% | 23 |
| 9 | CHAN, S , CHOI, EA , SHI, YS , (2011) PRE-MRNA 3 '-END PROCESSING COMPLEX ASSEMBLY AND FUNCTION.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 2. ISSUE 3. P. 321-335 | 81 | 73% | 43 |
| 10 | TIAN, B , MANLEY, JL , (2013) ALTERNATIVE CLEAVAGE AND POLYADENYLATION: THE LONG AND SHORT OF IT.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 38. ISSUE 6. P. 312 -320 | 67 | 65% | 84 |
Classes with closest relation at Level 1 |