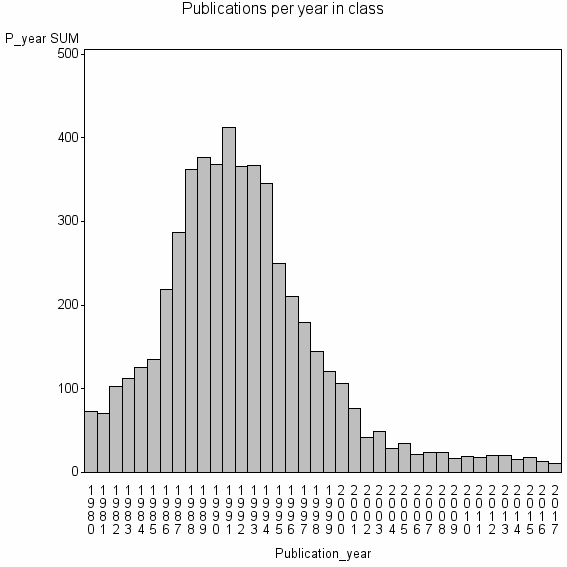
Class information for: |
Basic class information |

Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 17 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//MICROBIOLOGY//JOURNAL OF BACTERIOLOGY | 725599 |
| 226 | 3 | GENETICS & HEREDITY//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 49202 |
| 2203 | 2 | GENOMICS//MEIOTIC DRIVE//MAMMALIAN GENOME | 5174 |
| 5815 | 1 | GENOMICS//YEAST ARTIFICIAL CHROMOSOME//TAR CLONING | 1600 |
| 8676 | 1 | CYTOGENETICS AND CELL GENETICS//NUCLEIC ACIDS RESEARCH//MRC HUMAN ECOGENET UNIT | 1260 |
| 13834 | 1 | MEIOTIC DRIVE//T HAPLOTYPE//T COMPLEX | 840 |
| 15809 | 1 | MAMMALIAN GENOME//GENOMICS//FREDERICK CANC ILBIONET INC | 717 |
| 22636 | 1 | NOTI LINKING CLONES//NOTI LINKING AND JUMPING CLONES//NOTL LINKING CLONES | 396 |
| 28460 | 1 | EPSCOR//ALEITHIA//BUCANDIN | 225 |
| 33589 | 1 | 439//AT RICH PRIMERS//DEOXYRIBO NUCLEIC ACID IMAGES | 136 |
Terms with highest relevance score |
| rank | Category | termType | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass |
|---|---|---|---|---|---|---|
| 1 | GENOMICS | journal | 283225 | 12% | 8% | 621 |
| 2 | MEIOTIC DRIVE | authKW | 189414 | 1% | 45% | 69 |
| 3 | MAMMALIAN GENOME | journal | 176015 | 6% | 9% | 320 |
| 4 | CYTOGENETICS AND CELL GENETICS | journal | 156620 | 6% | 9% | 295 |
| 5 | NUCLEIC ACIDS RESEARCH | journal | 116538 | 17% | 2% | 881 |
| 6 | GENETICS & HEREDITY | WoSSC | 103092 | 58% | 1% | 3000 |
| 7 | T HAPLOTYPE | authKW | 86602 | 0% | 89% | 16 |
| 8 | T COMPLEX | authKW | 53660 | 0% | 38% | 23 |
| 9 | YEAST ARTIFICIAL CHROMOSOME | authKW | 45259 | 1% | 28% | 27 |
| 10 | SOMATIC CELL AND MOLECULAR GENETICS | journal | 44000 | 2% | 9% | 81 |
Web of Science journal categories |
| chi_square_rank | Category | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 103092 | 58% | 1% | 3000 |
| 2 | Biotechnology & Applied Microbiology | 13842 | 23% | 0% | 1176 |
| 3 | Biochemistry & Molecular Biology | 12908 | 42% | 0% | 2194 |
| 4 | Cell Biology | 2123 | 12% | 0% | 629 |
| 5 | Biochemical Research Methods | 805 | 5% | 0% | 263 |
| 6 | Evolutionary Biology | 587 | 3% | 0% | 136 |
| 7 | Biology | 303 | 3% | 0% | 149 |
| 8 | Industrial Relations & Labor | 176 | 1% | 0% | 29 |
| 9 | Developmental Biology | 127 | 1% | 0% | 71 |
| 10 | Mathematical & Computational Biology | 101 | 1% | 0% | 59 |
Address terms |
| chi_square_rank | term | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass |
|---|---|---|---|---|---|
| 1 | MRC HUMAN ECOGENET UNIT | 24358 | 0% | 100% | 4 |
| 2 | CNRS URA 1335 | 12681 | 0% | 42% | 5 |
| 3 | FREDERICK CANC ILBIONET INC | 12179 | 0% | 100% | 2 |
| 4 | INSERM U184 11 RUE HUMANN | 12179 | 0% | 100% | 2 |
| 5 | LGME CNRS | 12179 | 0% | 100% | 2 |
| 6 | MED GENET MED MOLEC MICROBIOL | 12179 | 0% | 100% | 2 |
| 7 | DEV TEAM MAMMALIAN CELLULAR DYNAM | 8118 | 0% | 67% | 2 |
| 8 | BIOL GENET EVOLUCAO | 7826 | 0% | 43% | 3 |
| 9 | SRSNR | 7826 | 0% | 43% | 3 |
| 10 | 439 | 6089 | 0% | 100% | 1 |
Journals |
| chi_square_rank | term | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass |
|---|---|---|---|---|---|
| 1 | GENOMICS | 283225 | 12% | 8% | 621 |
| 2 | MAMMALIAN GENOME | 176015 | 6% | 9% | 320 |
| 3 | CYTOGENETICS AND CELL GENETICS | 156620 | 6% | 9% | 295 |
| 4 | NUCLEIC ACIDS RESEARCH | 116538 | 17% | 2% | 881 |
| 5 | SOMATIC CELL AND MOLECULAR GENETICS | 44000 | 2% | 9% | 81 |
| 6 | SOMATIC CELL GENETICS | 27056 | 1% | 14% | 32 |
| 7 | GENETIC ANALYSIS-BIOMOLECULAR ENGINEERING | 23124 | 1% | 12% | 32 |
| 8 | GENETICAL RESEARCH | 18655 | 1% | 6% | 50 |
| 9 | ANNALS OF HUMAN GENETICS | 14122 | 1% | 4% | 64 |
| 10 | GENETICS | 10017 | 3% | 1% | 148 |
Author Key Words |
| chi_square_rank | term | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass | LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MEIOTIC DRIVE | 189414 | 1% | 45% | 69 | Search MEIOTIC+DRIVE | Search MEIOTIC+DRIVE |
| 2 | T HAPLOTYPE | 86602 | 0% | 89% | 16 | Search T+HAPLOTYPE | Search T+HAPLOTYPE |
| 3 | T COMPLEX | 53660 | 0% | 38% | 23 | Search T+COMPLEX | Search T+COMPLEX |
| 4 | YEAST ARTIFICIAL CHROMOSOME | 45259 | 1% | 28% | 27 | Search YEAST+ARTIFICIAL+CHROMOSOME | Search YEAST+ARTIFICIAL+CHROMOSOME |
| 5 | TAR CLONING | 22946 | 0% | 54% | 7 | Search TAR+CLONING | Search TAR+CLONING |
| 6 | TRANSMISSION DISTORTION | 21643 | 0% | 44% | 8 | Search TRANSMISSION+DISTORTION | Search TRANSMISSION+DISTORTION |
| 7 | TRANSMISSION RATIO DISTORTION | 20375 | 0% | 28% | 12 | Search TRANSMISSION+RATIO+DISTORTION | Search TRANSMISSION+RATIO+DISTORTION |
| 8 | DDK SYNDROME | 19485 | 0% | 80% | 4 | Search DDK+SYNDROME | Search DDK+SYNDROME |
| 9 | NOTI LINKING CLONES | 19026 | 0% | 63% | 5 | Search NOTI+LINKING+CLONES | Search NOTI+LINKING+CLONES |
| 10 | CLONE ORDERING | 18268 | 0% | 100% | 3 | Search CLONE+ORDERING | Search CLONE+ORDERING |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
Classes with closest relation at Level 2 |