Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
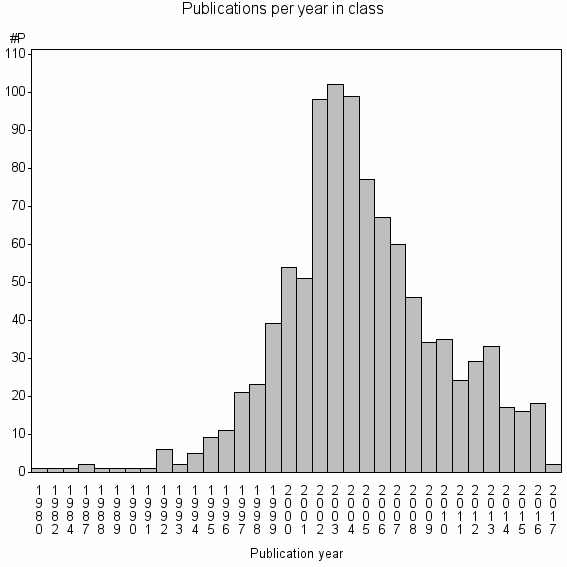
| 11368 | 987 | 35.3 | 87% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 2179 | 2 | REFERENCE GENES//GENORM//NORMFINDER | 4941 |
| 11368 | 1 | RNA AMPLIFICATION//LASER CAPTURE MICRODISSECTION//LASER MICRODISSECTION | 987 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RNA AMPLIFICATION | authKW | 667917 | 5% | 47% | 46 |
| 2 | LASER CAPTURE MICRODISSECTION | authKW | 604516 | 12% | 16% | 121 |
| 3 | LASER MICRODISSECTION | authKW | 227044 | 6% | 13% | 55 |
| 4 | PATHOGENET UNIT | address | 192122 | 2% | 30% | 21 |
| 5 | LASER PRESSURE CATAPULTING | authKW | 164987 | 1% | 67% | 8 |
| 6 | EXPRESSION MICRODISSECTION | authKW | 123744 | 0% | 100% | 4 |
| 7 | LINEAR RNA AMPLIFICATION | authKW | 123744 | 0% | 100% | 4 |
| 8 | MICROPALLET ARRAYS | authKW | 123744 | 0% | 100% | 4 |
| 9 | CANC GENOME ANAT PROJECT | address | 116457 | 1% | 47% | 8 |
| 10 | VOXELATION | authKW | 110483 | 1% | 71% | 5 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemical Research Methods | 4691 | 23% | 0% | 231 |
| 2 | Pathology | 2770 | 15% | 0% | 152 |
| 3 | Biochemistry & Molecular Biology | 696 | 25% | 0% | 249 |
| 4 | Genetics & Heredity | 481 | 10% | 0% | 103 |
| 5 | Biotechnology & Applied Microbiology | 448 | 10% | 0% | 102 |
| 6 | Neurosciences | 350 | 13% | 0% | 131 |
| 7 | Cell Biology | 229 | 10% | 0% | 96 |
| 8 | Oncology | 132 | 8% | 0% | 80 |
| 9 | Chemistry, Analytical | 92 | 6% | 0% | 56 |
| 10 | Medicine, Research & Experimental | 80 | 5% | 0% | 45 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PATHOGENET UNIT | 192122 | 2% | 30% | 21 |
| 2 | CANC GENOME ANAT PROJECT | 116457 | 1% | 47% | 8 |
| 3 | MURINE MICROARRAY AFFYMETRIX IL | 61872 | 0% | 100% | 2 |
| 4 | RUMENTAT DEV OURCE BIOENGN PHYS SC | 61872 | 0% | 100% | 2 |
| 5 | HORMONE RECEPTOR | 59486 | 1% | 38% | 5 |
| 6 | LASER MOL BIOL | 55682 | 0% | 60% | 3 |
| 7 | NATHAN KLINE | 52259 | 1% | 13% | 13 |
| 8 | NATHAN KLINE DEMENTIA | 41247 | 0% | 67% | 2 |
| 9 | TISSUE PROTEOM UNIT | 35350 | 0% | 29% | 4 |
| 10 | INTEGRATED NANOSYST IL | 33544 | 1% | 14% | 8 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOTECHNIQUES | 13228 | 5% | 1% | 51 |
| 2 | DIAGNOSTIC MOLECULAR PATHOLOGY | 3946 | 1% | 1% | 10 |
| 3 | JOURNAL OF MOLECULAR DIAGNOSTICS | 3887 | 1% | 1% | 12 |
| 4 | JOURNAL OF CLINICAL PATHOLOGY-MOLECULAR PATHOLOGY | 3702 | 1% | 2% | 7 |
| 5 | ARCHAOLOGISCHES NACHRICHTENBLATT | 3390 | 0% | 4% | 3 |
| 6 | MUTATION RESEARCH-GENOMICS | 3169 | 0% | 5% | 2 |
| 7 | JOURNAL OF NEUROSCIENCE METHODS | 2589 | 3% | 0% | 25 |
| 8 | BRAIN RESEARCH PROTOCOLS | 2270 | 1% | 1% | 6 |
| 9 | GENETIC ENGINEERING NEWS | 2250 | 1% | 1% | 8 |
| 10 | BMC GENOMICS | 2146 | 3% | 0% | 26 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA AMPLIFICATION | 667917 | 5% | 47% | 46 | Search RNA+AMPLIFICATION | Search RNA+AMPLIFICATION |
| 2 | LASER CAPTURE MICRODISSECTION | 604516 | 12% | 16% | 121 | Search LASER+CAPTURE+MICRODISSECTION | Search LASER+CAPTURE+MICRODISSECTION |
| 3 | LASER MICRODISSECTION | 227044 | 6% | 13% | 55 | Search LASER+MICRODISSECTION | Search LASER+MICRODISSECTION |
| 4 | LASER PRESSURE CATAPULTING | 164987 | 1% | 67% | 8 | Search LASER+PRESSURE+CATAPULTING | Search LASER+PRESSURE+CATAPULTING |
| 5 | EXPRESSION MICRODISSECTION | 123744 | 0% | 100% | 4 | Search EXPRESSION+MICRODISSECTION | Search EXPRESSION+MICRODISSECTION |
| 6 | LINEAR RNA AMPLIFICATION | 123744 | 0% | 100% | 4 | Search LINEAR+RNA+AMPLIFICATION | Search LINEAR+RNA+AMPLIFICATION |
| 7 | MICROPALLET ARRAYS | 123744 | 0% | 100% | 4 | Search MICROPALLET+ARRAYS | Search MICROPALLET+ARRAYS |
| 8 | VOXELATION | 110483 | 1% | 71% | 5 | Search VOXELATION | Search VOXELATION |
| 9 | MICRODISSECTION | 98107 | 5% | 7% | 45 | Search MICRODISSECTION | Search MICRODISSECTION |
| 10 | CRNA TARGET | 92808 | 0% | 100% | 3 | Search CRNA+TARGET | Search CRNA+TARGET |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | DATTA, S , MALHOTRA, L , DICKERSON, R , CHAFFEE, S , SEN, CK , ROY, S , (2015) LASER CAPTURE MICRODISSECTION: BIG DATA FROM SMALL SAMPLES.HISTOLOGY AND HISTOPATHOLOGY. VOL. 30. ISSUE 11. P. 1255 -1269 | 49 | 54% | 11 |
| 2 | NYGAARD, V , HOVIG, E , (2006) OPTIONS AVAILABLE FOR PROFILING SMALL SAMPLES: A REVIEW OF SAMPLE AMPLIFICATION TECHNOLOGY WHEN COMBINED WITH MICROARRAY PROFILING.NUCLEIC ACIDS RESEARCH. VOL. 34. ISSUE 3. P. 996-1014 | 54 | 59% | 81 |
| 3 | GINSBERG, SD , (2005) RNA AMPLIFICATION STRATEGIES FOR SMALL SAMPLE POPULATIONS.METHODS. VOL. 37. ISSUE 3. P. 229-237 | 36 | 82% | 40 |
| 4 | NELSON, T , TAUSTA, SL , GANDOTRA, N , LIU, T , (2006) LASER MICRODISSECTION OF PLANT TISSUE: WHAT YOU SEE IS WHAT YOU GET.ANNUAL REVIEW OF PLANT BIOLOGY. VOL. 57. ISSUE . P. 181 -201 | 51 | 52% | 79 |
| 5 | PEANO, C , SEVERGNINI, M , CIFOLA, I , DE BELLIS, G , BATTAGLIA, C , (2006) TRANSCRIPTOME AMPLIFICATION METHODS IN GENE EXPRESSION PROFILING.EXPERT REVIEW OF MOLECULAR DIAGNOSTICS. VOL. 6. ISSUE 3. P. 465-480 | 42 | 68% | 14 |
| 6 | KAWASAKI, ES , (2004) MICROARRAYS AND THE GENE EXPRESSION PROFILE OF A SINGLE CELL.APPLICATIONS OF BIOINFORMATICS IN CANCER DETECTION. VOL. 1020. ISSUE . P. 92 -100 | 40 | 74% | 21 |
| 7 | CLEMENT-ZIZA, M , GENTIEN, D , LYONNET, S , THIERY, JP , BESMOND, C , DECRAENE, C , (2009) EVALUATION OF METHODS FOR AMPLIFICATION OF PICOGRAM AMOUNTS OF TOTAL RNA FOR WHOLE GENOME EXPRESSION PROFILING.BMC GENOMICS. VOL. 10. ISSUE . P. - | 30 | 73% | 31 |
| 8 | ALLDRED, MJ , CHE, S , GINSBERG, SD , (2008) TERMINAL CONTINUATION (TC) RNA AMPLIFICATION ENABLES EXPRESSION PROFILING USING MINUTE RNA INPUT OBTAINED FROM MOUSE BRAIN.INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. VOL. 9. ISSUE 11. P. 2091 -2104 | 27 | 84% | 3 |
| 9 | GROVER, PK , CUMMINS, AG , PRICE, TJ , ROBERTS-THOMSON, IC , HARDINGHAM, JE , (2012) A SIMPLE, COST-EFFECTIVE AND FLEXIBLE METHOD FOR PROCESSING OF SNAP-FROZEN TISSUE TO PREPARE LARGE AMOUNTS OF INTACT RNA USING LASER MICRODISSECTION.BIOCHIMIE. VOL. 94. ISSUE 12. P. 2491-2497 | 31 | 63% | 2 |
| 10 | FEND, F , KREMER, M , QUINTANILLA-MARTINEZ, L , (2000) LASER CAPTURE MICRODISSECTION: METHODICAL ASPECTS AND APPLICATIONS WITH EMPHASIS ON IMMUNO-LASER CAPTURE MICRODISSECTION.PATHOBIOLOGY. VOL. 68. ISSUE 4-5. P. 209 -214 | 33 | 73% | 33 |
Classes with closest relation at Level 1 |