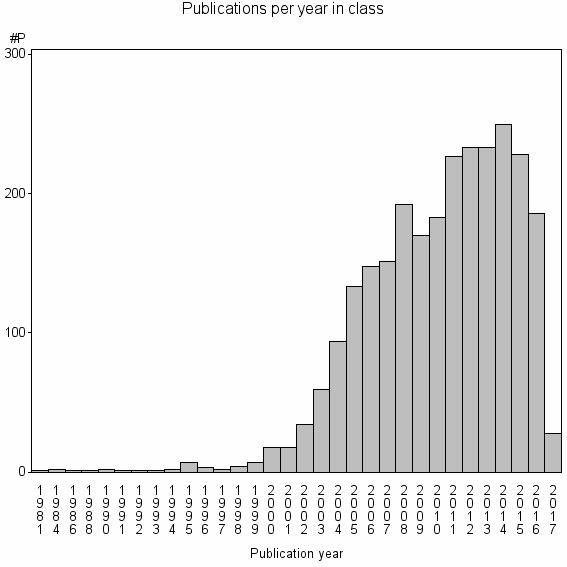
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 1508 | 2620 | 37.8 | 86% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 157 | 3 | JOURNAL OF THE AMERICAN SOCIETY FOR MASS SPECTROMETRY//BIOCHEMICAL RESEARCH METHODS//INTERNATIONAL JOURNAL OF MASS SPECTROMETRY | 61558 |
| 370 | 2 | PROTEOMICS//JOURNAL OF PROTEOME RESEARCH//BIOCHEMICAL RESEARCH METHODS | 17272 |
| 1508 | 1 | PEPTIDE IDENTIFICATION//JOURNAL OF PROTEOME RESEARCH//PROTEIN IDENTIFICATION | 2620 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PEPTIDE IDENTIFICATION | authKW | 807943 | 4% | 67% | 103 |
| 2 | JOURNAL OF PROTEOME RESEARCH | journal | 505833 | 20% | 8% | 514 |
| 3 | PROTEIN IDENTIFICATION | authKW | 453990 | 5% | 27% | 144 |
| 4 | PROTEOGENOMICS | authKW | 424262 | 2% | 58% | 63 |
| 5 | MISSING PROTEINS | authKW | 339864 | 1% | 83% | 35 |
| 6 | PROTEIN INFERENCE | authKW | 291322 | 1% | 100% | 25 |
| 7 | PROTEOMICS | journal | 259884 | 14% | 6% | 357 |
| 8 | HUMAN PROTEOME PROJECT | authKW | 249688 | 1% | 71% | 30 |
| 9 | PROTEOMICS STANDARDS INITIATIVE | authKW | 221405 | 1% | 100% | 19 |
| 10 | SHOTGUN PROTEOMICS | authKW | 203686 | 3% | 23% | 77 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemical Research Methods | 125945 | 72% | 1% | 1890 |
| 2 | Mathematical & Computational Biology | 12472 | 13% | 0% | 337 |
| 3 | Biotechnology & Applied Microbiology | 3422 | 16% | 0% | 429 |
| 4 | Chemistry, Analytical | 3296 | 16% | 0% | 427 |
| 5 | Spectroscopy | 1585 | 7% | 0% | 192 |
| 6 | Biochemistry & Molecular Biology | 1224 | 22% | 0% | 565 |
| 7 | Genetics & Heredity | 101 | 4% | 0% | 107 |
| 8 | Statistics & Probability | 35 | 1% | 0% | 38 |
| 9 | Computer Science, Interdisciplinary Applications | 31 | 2% | 0% | 42 |
| 10 | Mathematics, Interdisciplinary Applications | 3 | 1% | 0% | 19 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROTEOME INFORMAT GRP | 117728 | 1% | 42% | 24 |
| 2 | EMBL OUTSTN | 116656 | 2% | 21% | 48 |
| 3 | MOL SYST BIOL | 94669 | 4% | 9% | 92 |
| 4 | MED PROT | 90365 | 3% | 12% | 66 |
| 5 | JIM AYERS PRECANC DETECT DIAG | 76472 | 1% | 41% | 16 |
| 6 | SYST ENGN COMP SCI PROGRAM | 60859 | 1% | 33% | 16 |
| 7 | STRUCT BIOL BIOINFORMAT PROGRAMME | 58264 | 0% | 100% | 5 |
| 8 | PROTE SIGNAL TRANSDUCT | 56199 | 1% | 12% | 39 |
| 9 | COMPUTAT MASS SPE OMETRY | 55480 | 0% | 48% | 10 |
| 10 | BEIJING PROTEOME | 50312 | 1% | 11% | 38 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF PROTEOME RESEARCH | 505833 | 20% | 8% | 514 |
| 2 | PROTEOMICS | 259884 | 14% | 6% | 357 |
| 3 | MOLECULAR & CELLULAR PROTEOMICS | 179690 | 8% | 7% | 218 |
| 4 | JOURNAL OF PROTEOMICS | 42365 | 4% | 4% | 98 |
| 5 | BIOINFORMATICS | 26802 | 6% | 1% | 157 |
| 6 | BMC BIOINFORMATICS | 16655 | 4% | 1% | 104 |
| 7 | JOURNAL OF THE AMERICAN SOCIETY FOR MASS SPECTROMETRY | 15954 | 3% | 2% | 80 |
| 8 | EXPERT REVIEW OF PROTEOMICS | 14270 | 1% | 4% | 30 |
| 9 | PROTEOME SCIENCE | 12048 | 1% | 4% | 24 |
| 10 | OMICS-A JOURNAL OF INTEGRATIVE BIOLOGY | 10341 | 1% | 4% | 25 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PEPTIDE IDENTIFICATION | 807943 | 4% | 67% | 103 | Search PEPTIDE+IDENTIFICATION | Search PEPTIDE+IDENTIFICATION |
| 2 | PROTEIN IDENTIFICATION | 453990 | 5% | 27% | 144 | Search PROTEIN+IDENTIFICATION | Search PROTEIN+IDENTIFICATION |
| 3 | PROTEOGENOMICS | 424262 | 2% | 58% | 63 | Search PROTEOGENOMICS | Search PROTEOGENOMICS |
| 4 | MISSING PROTEINS | 339864 | 1% | 83% | 35 | Search MISSING+PROTEINS | Search MISSING+PROTEINS |
| 5 | PROTEIN INFERENCE | 291322 | 1% | 100% | 25 | Search PROTEIN+INFERENCE | Search PROTEIN+INFERENCE |
| 6 | HUMAN PROTEOME PROJECT | 249688 | 1% | 71% | 30 | Search HUMAN+PROTEOME+PROJECT | Search HUMAN+PROTEOME+PROJECT |
| 7 | PROTEOMICS STANDARDS INITIATIVE | 221405 | 1% | 100% | 19 | Search PROTEOMICS+STANDARDS+INITIATIVE | Search PROTEOMICS+STANDARDS+INITIATIVE |
| 8 | SHOTGUN PROTEOMICS | 203686 | 3% | 23% | 77 | Search SHOTGUN+PROTEOMICS | Search SHOTGUN+PROTEOMICS |
| 9 | DATABASE SEARCH | 187719 | 3% | 23% | 70 | Search DATABASE+SEARCH | Search DATABASE+SEARCH |
| 10 | BIOINFORMATICS | 179775 | 12% | 5% | 302 | Search BIOINFORMATICS | Search BIOINFORMATICS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | NESVIZHSKII, AI , (2010) A SURVEY OF COMPUTATIONAL METHODS AND ERROR RATE ESTIMATION PROCEDURES FOR PEPTIDE AND PROTEIN IDENTIFICATION IN SHOTGUN PROTEOMICS.JOURNAL OF PROTEOMICS. VOL. 73. ISSUE 11. P. 2092 -2123 | 213 | 82% | 171 |
| 2 | VIZCAINO, JA , CSORDAS, A , DEL-TORO, N , DIANES, JA , GRISS, J , LAVIDAS, I , MAYER, G , PEREZ-RIVEROL, Y , REISINGER, F , TERNENT, T , ET AL (2016) 2016 UPDATE OF THE PRIDE DATABASE AND ITS RELATED TOOLS.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE D1. P. D447 -D456 | 36 | 75% | 217 |
| 3 | COX, J , NEUHAUSER, N , MICHALSKI, A , SCHELTEMA, RA , OLSEN, JV , MANN, M , (2011) ANDROMEDA: A PEPTIDE SEARCH ENGINE INTEGRATED INTO THE MAXQUANT ENVIRONMENT.JOURNAL OF PROTEOME RESEARCH. VOL. 10. ISSUE 4. P. 1794 -1805 | 48 | 79% | 935 |
| 4 | ZHANG, YY , FONSLOW, BR , SHAN, B , BAEK, MC , YATES, JR , (2013) PROTEIN ANALYSIS BY SHOTGUN/BOTTOM-UP PROTEOMICS.CHEMICAL REVIEWS. VOL. 113. ISSUE 4. P. 2343 -2394 | 185 | 24% | 214 |
| 5 | NESVIZHSKII, AI , VITEK, O , AEBERSOLD, R , (2007) ANALYSIS AND VALIDATION OF PROTEOMIC DATA GENERATED BY TANDEM MASS SPECTROMETRY.NATURE METHODS. VOL. 4. ISSUE 10. P. 787-797 | 82 | 78% | 310 |
| 6 | PEREZ-RIVEROL, Y , ALPI, E , WANG, R , HERMJAKOB, H , VIZCAINO, JA , (2015) MAKING PROTEOMICS DATA ACCESSIBLE AND REUSABLE: CURRENT STATE OF PROTEOMICS DATABASES AND REPOSITORIES.PROTEOMICS. VOL. 15. ISSUE 5-6. P. 930 -949 | 79 | 73% | 22 |
| 7 | PEREZ-RIVEROL, Y , WANG, R , HERMJAKOB, H , MULLER, M , VESADA, V , VIZCAINO, JA , (2014) OPEN SOURCE LIBRARIES AND FRAMEWORKS FOR MASS SPECTROMETRY BASED PROTEOMICS: A DEVELOPER'S PERSPECTIVE.BIOCHIMICA ET BIOPHYSICA ACTA-PROTEINS AND PROTEOMICS. VOL. 1844. ISSUE 1. P. 63 -76 | 95 | 69% | 14 |
| 8 | BLEIN-NICOLAS, M , ZIVY, M , (2016) THOUSAND AND ONE WAYS TO QUANTIFY AND COMPARE PROTEIN ABUNDANCES IN LABEL-FREE BOTTOM-UP PROTEOMICS.BIOCHIMICA ET BIOPHYSICA ACTA-PROTEINS AND PROTEOMICS. VOL. 1864. ISSUE 8. P. 883 -895 | 73 | 78% | 2 |
| 9 | FENG, XD , MA, J , CHANG, C , BAI, MZ , ZHU, YP , SHU, KX , (2016) THE APPLICATION AND PROGRESS OF TARGET-DECOY DATABASE SEARCH STRATEGY IN IDENTIFICATION AND QUALITY CONTROL OF TANDEM MASS SPECTROMETRY DATA IN SHOTGUN PROTEOMICS.PROGRESS IN BIOCHEMISTRY AND BIOPHYSICS. VOL. 43. ISSUE 7. P. 661 -672 | 64 | 98% | 0 |
| 10 | FORNER, F , FOSTER, LJ , TOPPO, S , (2007) MASS SPECTROMETRY DATA ANALYSIS IN THE PROTEOMICS ERA.CURRENT BIOINFORMATICS. VOL. 2. ISSUE 1. P. 63 -93 | 128 | 57% | 17 |
Classes with closest relation at Level 1 |