Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
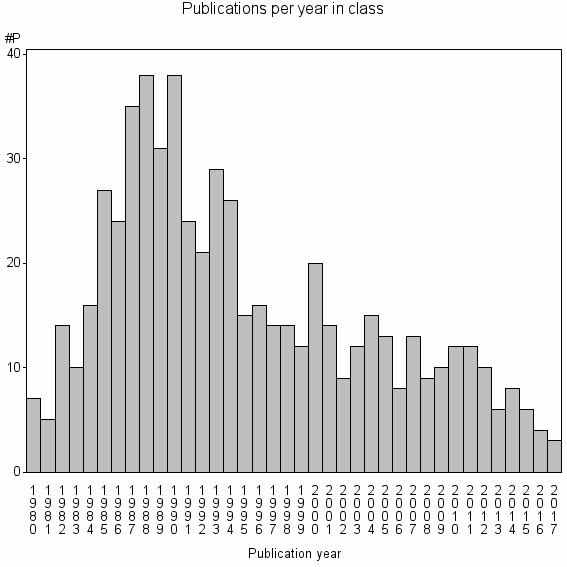
| 17419 | 600 | 40.7 | 75% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 71 | 3 | GENETICS & HEREDITY//CHROMATIN//CELL BIOLOGY | 83302 |
| 225 | 2 | YEAST//SACCHAROMYCES CEREVISIAE//SCHIZOSACCHAROMYCES POMBE | 20517 |
| 17419 | 1 | INFORMAT GENET DEV//BCY1//CDC25P | 600 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | INFORMAT GENET DEV | address | 207799 | 1% | 58% | 7 |
| 2 | BCY1 | authKW | 178112 | 1% | 50% | 7 |
| 3 | CDC25P | authKW | 152673 | 1% | 100% | 3 |
| 4 | RAS2P | authKW | 152673 | 1% | 100% | 3 |
| 5 | MSN2P | authKW | 152667 | 1% | 50% | 6 |
| 6 | FITNESS BASED INTERFERENTIAL GENETICS | authKW | 101782 | 0% | 100% | 2 |
| 7 | RAS2 GTP | authKW | 101782 | 0% | 100% | 2 |
| 8 | RAS CAMP PATHWAY | authKW | 97861 | 1% | 38% | 5 |
| 9 | GPA2 | authKW | 90469 | 1% | 44% | 4 |
| 10 | MOL CELBIOL | address | 86067 | 3% | 11% | 15 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 3554 | 63% | 0% | 376 |
| 2 | Cell Biology | 2248 | 32% | 0% | 192 |
| 3 | Microbiology | 1416 | 21% | 0% | 127 |
| 4 | Mycology | 1263 | 6% | 0% | 36 |
| 5 | Genetics & Heredity | 656 | 15% | 0% | 89 |
| 6 | Biophysics | 435 | 11% | 0% | 64 |
| 7 | Biotechnology & Applied Microbiology | 117 | 7% | 0% | 44 |
| 8 | Biology | 15 | 2% | 0% | 13 |
| 9 | Biochemical Research Methods | 9 | 2% | 0% | 14 |
| 10 | CYTOLOGY & HISTOLOGY | 5 | 0% | 0% | 1 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | INFORMAT GENET DEV | 207799 | 1% | 58% | 7 |
| 2 | MOL CELBIOL | 86067 | 3% | 11% | 15 |
| 3 | BIOTECHNOL K681304 | 50891 | 0% | 100% | 1 |
| 4 | BOT MIKROBIOLFLANDERS INTERUNIVERS B | 50891 | 0% | 100% | 1 |
| 5 | CNRS UPS UMR 8621 | 50891 | 0% | 100% | 1 |
| 6 | CNRSUMR 2225 | 50891 | 0% | 100% | 1 |
| 7 | COMISSAO NACL DESENVOLVIMENTO TECNOL NUCL | 50891 | 0% | 100% | 1 |
| 8 | CORP MFG SYST | 50891 | 0% | 100% | 1 |
| 9 | DEV MO BIOL | 50891 | 0% | 100% | 1 |
| 10 | DIPARTIMENHTO BIOTECNOL BIOSCI | 50891 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | YEAST | 8660 | 4% | 1% | 22 |
| 2 | CURRENT GENETICS | 7944 | 4% | 1% | 24 |
| 3 | MOLECULAR AND CELLULAR BIOLOGY | 3996 | 7% | 0% | 42 |
| 4 | MOLECULAR AND GENERAL GENETICS | 2362 | 3% | 0% | 15 |
| 5 | JOURNAL OF GENERAL MICROBIOLOGY | 1876 | 2% | 0% | 14 |
| 6 | MICROBIOLOGY-SGM | 1439 | 3% | 0% | 15 |
| 7 | MOLECULAR GENETICS AND GENOMICS | 1249 | 1% | 0% | 7 |
| 8 | FEBS LETTERS | 1015 | 5% | 0% | 29 |
| 9 | EMBO JOURNAL | 938 | 3% | 0% | 18 |
| 10 | EUROPEAN JOURNAL OF BIOCHEMISTRY | 902 | 3% | 0% | 19 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | BCY1 | 178112 | 1% | 50% | 7 | Search BCY1 | Search BCY1 |
| 2 | CDC25P | 152673 | 1% | 100% | 3 | Search CDC25P | Search CDC25P |
| 3 | RAS2P | 152673 | 1% | 100% | 3 | Search RAS2P | Search RAS2P |
| 4 | MSN2P | 152667 | 1% | 50% | 6 | Search MSN2P | Search MSN2P |
| 5 | FITNESS BASED INTERFERENTIAL GENETICS | 101782 | 0% | 100% | 2 | Search FITNESS+BASED+INTERFERENTIAL+GENETICS | Search FITNESS+BASED+INTERFERENTIAL+GENETICS |
| 6 | RAS2 GTP | 101782 | 0% | 100% | 2 | Search RAS2+GTP | Search RAS2+GTP |
| 7 | RAS CAMP PATHWAY | 97861 | 1% | 38% | 5 | Search RAS+CAMP+PATHWAY | Search RAS+CAMP+PATHWAY |
| 8 | GPA2 | 90469 | 1% | 44% | 4 | Search GPA2 | Search GPA2 |
| 9 | STRE | 84812 | 1% | 33% | 5 | Search STRE | Search STRE |
| 10 | GSY2 | 67853 | 0% | 67% | 2 | Search GSY2 | Search GSY2 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | VANDAMME, J , CASTERMANS, D , THEVELEIN, JM , (2012) MOLECULAR MECHANISMS OF FEEDBACK INHIBITION OF PROTEIN KINASE A ON INTRACELLULAR CAMP ACCUMULATION.CELLULAR SIGNALLING. VOL. 24. ISSUE 8. P. 1610-1618 | 74 | 49% | 23 |
| 2 | THEVELEIN, JM , (1992) THE RAS-ADENYLATE CYCLASE PATHWAY AND CELL-CYCLE CONTROL IN SACCHAROMYCES-CEREVISIAE.ANTONIE VAN LEEUWENHOEK INTERNATIONAL JOURNAL OF GENERAL AND MOLECULAR MICROBIOLOGY. VOL. 62. ISSUE 1-2. P. 109-130 | 75 | 58% | 88 |
| 3 | THEVELEIN, JM , DE WINDE, JH , (1999) NOVEL SENSING MECHANISMS AND TARGETS FOR THE CAMP-PROTEIN KINASE A PATHWAY IN THE YEAST SACCHAROMYCES CEREVISIAE.MOLECULAR MICROBIOLOGY. VOL. 33. ISSUE 5. P. 904-918 | 47 | 50% | 396 |
| 4 | COLOMBO, S , MA, PS , CAUWENBERG, L , WINDERICKX, J , CRAUWELS, M , TEUNISSEN, A , NAUWELAERS, D , DE WINDE, JH , GORWA, MF , COLAVIZZA, D , ET AL (1998) INVOLVEMENT OF DISTINCT G-PROTEINS, GPA2 AND RAS, IN GLUCOSE- AND INTRACELLULAR ACIDIFICATION-INDUCED CAMP SIGNALLING IN THE YEAST SACCHAROMYCES CEREVISIAE.EMBO JOURNAL. VOL. 17. ISSUE 12. P. 3326-3341 | 49 | 59% | 174 |
| 5 | COLOMBO, S , PAIARDI, C , PARDONS, K , WINDERICKX, J , MARTEGANI, E , (2014) EVIDENCE FOR ADENYLATE CYCLASE AS A SCAFFOLD PROTEIN FOR RAS2-IRA INTERACTION IN SACCHAROMYCES CEREVISIE.CELLULAR SIGNALLING. VOL. 26. ISSUE 5. P. 1147 -1154 | 29 | 76% | 4 |
| 6 | BELOTTI, F , TISI, R , MARTEGANI, E , (2006) THE N-TERMINAL REGION OF THE SACCHAROMYCES CEREVISIAE RASGEF CDC25 IS REQUIRED FOR NUTRIENT-DEPENDENT CELL-SIZE REGULATION.MICROBIOLOGY-SGM. VOL. 152. ISSUE . P. 1231 -1242 | 38 | 70% | 9 |
| 7 | BAI, XJ , DONG, J , (2010) SERINE214 OF RAS2P PLAYS A ROLE IN THE FEEDBACK REGULATION OF THE RAS-CAMP PATHWAY IN THE YEAST SACCHAROMYCES CEREVISIAE.FEBS LETTERS. VOL. 584. ISSUE 11. P. 2333 -2338 | 28 | 85% | 0 |
| 8 | DONG, JA , BAI, XJ , (2011) THE MEMBRANE LOCALIZATION OF RAS2P AND THE ASSOCIATION BETWEEN CDC25P AND RAS2-GTP ARE REGULATED BY PROTEIN KINASE A (PKA) IN THE YEAST SACCHAROMYCES CEREVISIAE.FEBS LETTERS. VOL. 585. ISSUE 8. P. 1127 -1134 | 23 | 92% | 6 |
| 9 | COLOMBO, S , RONCHETTI, D , THEVELEIN, JM , WINDERICKX, J , MARTEGANI, E , (2004) ACTIVATION STATE OF THE RAS2 PROTEIN AND GLUCOSE-INDUCED SIGNALING IN SACCHAROMYCES CEREVISIAE.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 279. ISSUE 45. P. 46715-46722 | 27 | 75% | 66 |
| 10 | UNO, I , (1992) ROLE OF SIGNAL-TRANSDUCTION SYSTEMS IN CELL-PROLIFERATION IN YEAST.INTERNATIONAL REVIEW OF CYTOLOGY-A SURVEY OF CELL BIOLOGY. VOL. 139. ISSUE . P. 309-332 | 66 | 51% | 3 |
Classes with closest relation at Level 1 |
| Rank | Class id | link |
|---|---|---|
| 1 | 18452 | IME1//IME2//PROSPORE MEMBRANE |
| 2 | 24640 | TBRPDEB1//TARGET REPURPOSING//ANTIINFECT SCREENING CORE |
| 3 | 5250 | GLUCOSE REPRESSION//MIG1//MALTOSE PERMEASE |
| 4 | 8143 | TREHALOSE//TREHALASE//TREHALOSE 6 PHOSPHATE SYNTHASE |
| 5 | 4031 | RAS//RAS PROTEINS//ONCOGENIC RAS P21 |
| 6 | 5999 | PHEROMONE RESPONSE//FLO11//PSEUDOHYPHAL GROWTH |
| 7 | 9347 | CHRONOLOGICAL LIFESPAN//REPLICATIVE LIFESPAN//YEAST APOPTOSIS |
| 8 | 20729 | PHO85//P I TRANSPORTER//PHO84 |
| 9 | 17553 | FRUCTOSE 1 6 BISPHOSPHATASE//FBPASE//MUSCLE FBPASE |
| 10 | 12036 | START//MBF//SIC1 |