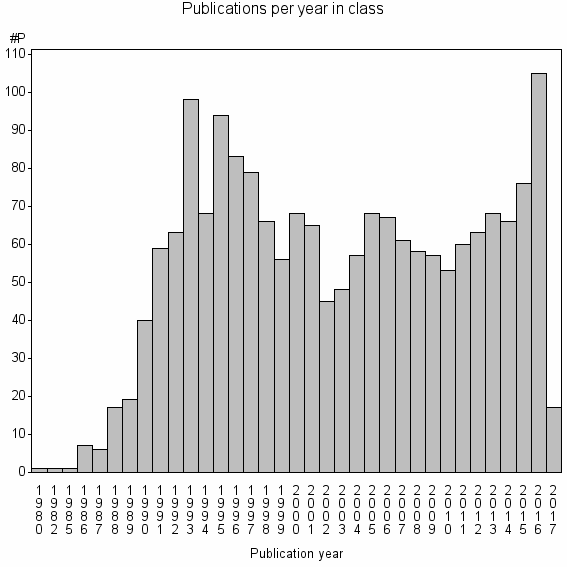
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 4031 | 1860 | 49.2 | 91% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 269 | 3 | CELL BIOLOGY//CAVEOLIN 1//CAVEOLAE | 44007 |
| 1911 | 2 | FARNESYLTRANSFERASE//RAS//RAL | 5888 |
| 4031 | 1 | RAS//RAS PROTEINS//ONCOGENIC RAS P21 | 1860 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RAS | authKW | 264793 | 13% | 7% | 233 |
| 2 | RAS PROTEINS | authKW | 198314 | 2% | 28% | 43 |
| 3 | ONCOGENIC RAS P21 | authKW | 180566 | 1% | 100% | 11 |
| 4 | CDC25MM | authKW | 131321 | 0% | 100% | 8 |
| 5 | K RAS4B | authKW | 117245 | 1% | 71% | 10 |
| 6 | RAS GRF | authKW | 109427 | 1% | 67% | 10 |
| 7 | ABT STRUKTURELLE BIOL | address | 94550 | 1% | 26% | 22 |
| 8 | GTP HYDROLYSIS | authKW | 90753 | 1% | 22% | 25 |
| 9 | EFFECTOR DOMAIN | authKW | 90272 | 1% | 50% | 11 |
| 10 | HSOS1 | authKW | 82076 | 0% | 100% | 5 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 11765 | 65% | 0% | 1201 |
| 2 | Cell Biology | 8985 | 36% | 0% | 670 |
| 3 | Biophysics | 1208 | 10% | 0% | 189 |
| 4 | Oncology | 926 | 13% | 0% | 249 |
| 5 | Genetics & Heredity | 415 | 8% | 0% | 141 |
| 6 | Biochemical Research Methods | 187 | 4% | 0% | 80 |
| 7 | Chemistry, Medicinal | 41 | 2% | 0% | 41 |
| 8 | Developmental Biology | 13 | 1% | 0% | 17 |
| 9 | Biology | 11 | 1% | 0% | 27 |
| 10 | Mathematical & Computational Biology | 2 | 1% | 0% | 10 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ABT STRUKTURELLE BIOL | 94550 | 1% | 26% | 22 |
| 2 | LEHRSTUHL BIOPHYS | 51553 | 1% | 14% | 22 |
| 3 | MAX PLANCK PARTNER | 49245 | 0% | 100% | 3 |
| 4 | FREDERICK CANC CANC INFLAMMAT PROG | 32830 | 0% | 100% | 2 |
| 5 | MOL STUDIES CELL REGULAT | 32830 | 0% | 100% | 2 |
| 6 | SIR ALASTAIR CURRIE CANC UK | 32830 | 0% | 100% | 2 |
| 7 | INTEGRAT BIOL PHARMACOL | 31411 | 2% | 5% | 39 |
| 8 | IBMCC CSIC USAL | 26259 | 0% | 40% | 4 |
| 9 | MED CELL BIOL PHARMACOL | 26259 | 0% | 40% | 4 |
| 10 | BIOPHYS PHYS BIOCHEM | 25684 | 1% | 6% | 27 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR AND CELLULAR BIOLOGY | 8329 | 6% | 0% | 107 |
| 2 | ONCOGENE | 5863 | 4% | 0% | 81 |
| 3 | JOURNAL OF BIOLOGICAL CHEMISTRY | 4560 | 11% | 0% | 210 |
| 4 | JOURNAL OF PROTEIN CHEMISTRY | 3527 | 1% | 1% | 18 |
| 5 | BIOCHEMISTRY | 2368 | 5% | 0% | 90 |
| 6 | STRUCTURE | 1528 | 1% | 1% | 19 |
| 7 | CELLULAR SIGNALLING | 1473 | 1% | 0% | 20 |
| 8 | CELL GROWTH & DIFFERENTIATION | 1391 | 1% | 1% | 11 |
| 9 | METHODS IN ENZYMOLOGY | 1107 | 2% | 0% | 34 |
| 10 | PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA | 1099 | 5% | 0% | 84 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RAS | 264793 | 13% | 7% | 233 | Search RAS | Search RAS |
| 2 | RAS PROTEINS | 198314 | 2% | 28% | 43 | Search RAS+PROTEINS | Search RAS+PROTEINS |
| 3 | ONCOGENIC RAS P21 | 180566 | 1% | 100% | 11 | Search ONCOGENIC+RAS+P21 | Search ONCOGENIC+RAS+P21 |
| 4 | CDC25MM | 131321 | 0% | 100% | 8 | Search CDC25MM | Search CDC25MM |
| 5 | K RAS4B | 117245 | 1% | 71% | 10 | Search K+RAS4B | Search K+RAS4B |
| 6 | RAS GRF | 109427 | 1% | 67% | 10 | Search RAS+GRF | Search RAS+GRF |
| 7 | GTP HYDROLYSIS | 90753 | 1% | 22% | 25 | Search GTP+HYDROLYSIS | Search GTP+HYDROLYSIS |
| 8 | EFFECTOR DOMAIN | 90272 | 1% | 50% | 11 | Search EFFECTOR+DOMAIN | Search EFFECTOR+DOMAIN |
| 9 | HSOS1 | 82076 | 0% | 100% | 5 | Search HSOS1 | Search HSOS1 |
| 10 | RASGRF1 | 81577 | 1% | 38% | 13 | Search RASGRF1 | Search RASGRF1 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LU, SY , JANG, H , MURATCIOGLU, S , GURSOY, A , KESKIN, O , NUSSINOV, R , ZHANG, J , (2016) RAS CONFORMATIONAL ENSEMBLES, ALLOSTERY, AND SIGNALING.CHEMICAL REVIEWS. VOL. 116. ISSUE 11. P. 6607 -6665 | 236 | 57% | 5 |
| 2 | PRAKASH, P , GORFE, AA , (2013) LESSONS FROM COMPUTER SIMULATIONS OF RAS PROTEINS IN SOLUTION AND IN MEMBRANE.BIOCHIMICA ET BIOPHYSICA ACTA-GENERAL SUBJECTS. VOL. 1830. ISSUE 11. P. 5211-5218 | 124 | 83% | 13 |
| 3 | LU, SY , JANG, H , GU, S , ZHANG, J , NUSSINOV, R , (2016) DRUGGING RAS GTPASE: A COMPREHENSIVE MECHANISTIC AND SIGNALING STRUCTURAL VIEW.CHEMICAL SOCIETY REVIEWS. VOL. 45. ISSUE 18. P. 4929 -4952 | 136 | 62% | 3 |
| 4 | MARCUS, K , MATTOS, C , (2015) DIRECT ATTACK ON RAS: INTRAMOLECULAR COMMUNICATION AND MUTATION-SPECIFIC EFFECTS.CLINICAL CANCER RESEARCH. VOL. 21. ISSUE 8. P. 1810 -1818 | 65 | 88% | 18 |
| 5 | PRAKASH, P , GORFE, AA , (2014) OVERVIEW OF SIMULATION STUDIES ON THE ENZYMATIC ACTIVITY AND CONFORMATIONAL DYNAMICS OF THE GTPASE RAS.MOLECULAR SIMULATION. VOL. 40. ISSUE 10-11. P. 839 -847 | 78 | 71% | 6 |
| 6 | BUDAY, L , DOWNWARD, J , (2008) MANY FACES OF RAS ACTIVATION.BIOCHIMICA ET BIOPHYSICA ACTA-REVIEWS ON CANCER. VOL. 1786. ISSUE 2. P. 178-187 | 92 | 61% | 83 |
| 7 | HENNIG, A , MARKWART, R , ESPARZA-FRANCO, MA , LADDS, G , RUBIO, I , (2015) RAS ACTIVATION REVISITED: ROLE OF GEF AND GAP SYSTEMS.BIOLOGICAL CHEMISTRY. VOL. 396. ISSUE 8. P. 831 -848 | 94 | 50% | 5 |
| 8 | SPIEGEL, J , CROMM, PM , ZIMMERMANN, G , GROSSMANN, TN , WALDMANN, H , (2014) SMALL-MOLECULE MODULATION OF RAS SIGNALING.NATURE CHEMICAL BIOLOGY. VOL. 10. ISSUE 8. P. 613 -622 | 60 | 60% | 41 |
| 9 | PRIOR, IA , HANCOCK, JF , (2012) RAS TRAFFICKING, LOCALIZATION AND COMPARTMENTALIZED SIGNALLING.SEMINARS IN CELL & DEVELOPMENTAL BIOLOGY. VOL. 23. ISSUE 2. P. 145-153 | 67 | 60% | 51 |
| 10 | HOBBS, GA , DER, CJ , ROSSMAN, KL , (2016) RAS ISOFORMS AND MUTATIONS IN CANCER AT A GLANCE.JOURNAL OF CELL SCIENCE. VOL. 129. ISSUE 7. P. 1287 -1292 | 46 | 65% | 6 |
Classes with closest relation at Level 1 |
| Rank | Class id | link |
|---|---|---|
| 1 | 33764 | FARNESYLTHIOSALICYLIC ACID//SALIRASIB//FTS |
| 2 | 14252 | RAL//EXOCYST//RLIP76 |
| 3 | 35582 | RASGAP//G3BP1//G3BP |
| 4 | 17419 | INFORMAT GENET DEV//BCY1//CDC25P |
| 5 | 11748 | RAF//LEO JENKINS CANC//KSR1 |
| 6 | 11178 | RAP1//EPAC//EPAC1 |
| 7 | 2802 | N RAS//RAS GENES//RAS ONCOGENE |
| 8 | 2877 | FARNESYLTRANSFERASE//FARNESYLTRANSFERASE INHIBITOR//PROTEIN FARNESYLTRANSFERASE |
| 9 | 1558 | RHO GTPASES//TIAM1//RHOB |
| 10 | 9970 | MAP KINASE KINASE//MAP KINASE//MAP KINASE ACTIVATOR |