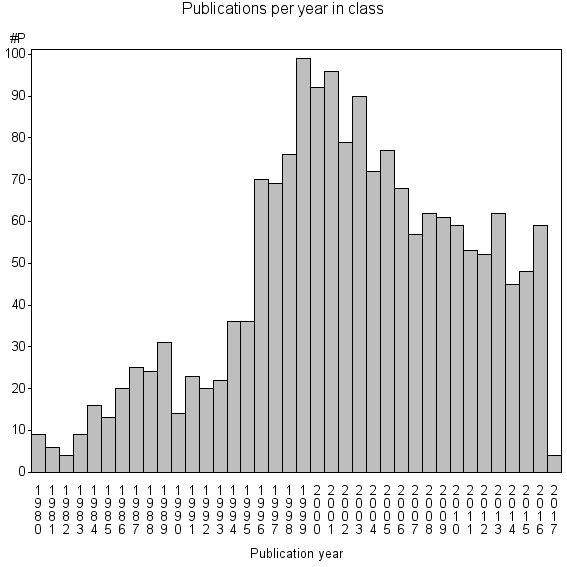
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 4599 | 1758 | 47.4 | 87% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 160 | 2 | WERNER SYNDROME//DNA POLYMERASE//DNA REPAIR | 22798 |
| 4599 | 1 | MISMATCH REPAIR//MUTS//DNA MISMATCH REPAIR | 1758 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MISMATCH REPAIR | authKW | 984279 | 13% | 25% | 229 |
| 2 | MUTS | authKW | 889881 | 4% | 70% | 73 |
| 3 | DNA MISMATCH REPAIR | authKW | 821387 | 8% | 35% | 136 |
| 4 | MUTL | authKW | 480271 | 2% | 71% | 39 |
| 5 | MSH2 | authKW | 162060 | 3% | 17% | 55 |
| 6 | MSH2 MSH6 | authKW | 156303 | 1% | 75% | 12 |
| 7 | MLH1 PMS1 | authKW | 138941 | 0% | 100% | 8 |
| 8 | MUTH | authKW | 117227 | 1% | 75% | 9 |
| 9 | METHYLATION TOLERANCE | authKW | 104206 | 0% | 100% | 6 |
| 10 | MUTS HOMOLOGUES | authKW | 104206 | 0% | 100% | 6 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 9017 | 30% | 0% | 532 |
| 2 | Biochemistry & Molecular Biology | 6440 | 50% | 0% | 887 |
| 3 | Toxicology | 2230 | 11% | 0% | 198 |
| 4 | Cell Biology | 2134 | 19% | 0% | 338 |
| 5 | Oncology | 1537 | 17% | 0% | 298 |
| 6 | Biotechnology & Applied Microbiology | 253 | 7% | 0% | 115 |
| 7 | Microbiology | 206 | 6% | 0% | 105 |
| 8 | Biophysics | 76 | 4% | 0% | 62 |
| 9 | Biology | 34 | 2% | 0% | 35 |
| 10 | Andrology | 15 | 0% | 0% | 4 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENET MOL BIOL PROGRAM | 61183 | 1% | 21% | 17 |
| 2 | BIOCHEM FB 08 | 60780 | 0% | 50% | 7 |
| 3 | SECT CHEM CARCINOGENESIS | 55575 | 0% | 80% | 4 |
| 4 | GENET BIOCHEM BRANCH | 49622 | 1% | 12% | 23 |
| 5 | ANORGAN ANALYT CHEM FB 08 | 34735 | 0% | 100% | 2 |
| 6 | DNA REPAIR MOL CARCINOGENESIS PROGRAM | 34735 | 0% | 100% | 2 |
| 7 | MED RADIOBIOL | 31282 | 1% | 11% | 16 |
| 8 | MOL CANC | 30943 | 2% | 5% | 33 |
| 9 | MED 0058 | 27130 | 0% | 31% | 5 |
| 10 | CHARLES A DANA HUMAN CANC GENET | 25257 | 0% | 36% | 4 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 73783 | 5% | 5% | 93 |
| 2 | GENETICS | 9499 | 5% | 1% | 84 |
| 3 | MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS | 8179 | 2% | 1% | 43 |
| 4 | MUTATION RESEARCH-DNA REPAIR | 7995 | 1% | 3% | 15 |
| 5 | NUCLEIC ACIDS RESEARCH | 5380 | 6% | 0% | 112 |
| 6 | JOURNAL OF BIOLOGICAL CHEMISTRY | 2925 | 9% | 0% | 165 |
| 7 | MOLECULAR AND CELLULAR BIOLOGY | 2365 | 3% | 0% | 56 |
| 8 | MOLECULAR CELL | 2276 | 2% | 0% | 27 |
| 9 | JOURNAL OF BACTERIOLOGY | 2049 | 4% | 0% | 64 |
| 10 | MOLECULAR AND GENERAL GENETICS | 2042 | 1% | 1% | 24 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MISMATCH REPAIR | 984279 | 13% | 25% | 229 | Search MISMATCH+REPAIR | Search MISMATCH+REPAIR |
| 2 | MUTS | 889881 | 4% | 70% | 73 | Search MUTS | Search MUTS |
| 3 | DNA MISMATCH REPAIR | 821387 | 8% | 35% | 136 | Search DNA+MISMATCH+REPAIR | Search DNA+MISMATCH+REPAIR |
| 4 | MUTL | 480271 | 2% | 71% | 39 | Search MUTL | Search MUTL |
| 5 | MSH2 | 162060 | 3% | 17% | 55 | Search MSH2 | Search MSH2 |
| 6 | MSH2 MSH6 | 156303 | 1% | 75% | 12 | Search MSH2+MSH6 | Search MSH2+MSH6 |
| 7 | MLH1 PMS1 | 138941 | 0% | 100% | 8 | Search MLH1+PMS1 | Search MLH1+PMS1 |
| 8 | MUTH | 117227 | 1% | 75% | 9 | Search MUTH | Search MUTH |
| 9 | METHYLATION TOLERANCE | 104206 | 0% | 100% | 6 | Search METHYLATION+TOLERANCE | Search METHYLATION+TOLERANCE |
| 10 | MUTS HOMOLOGUES | 104206 | 0% | 100% | 6 | Search MUTS+HOMOLOGUES | Search MUTS+HOMOLOGUES |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | IYER, RR , PLUCIENNIK, A , BURDETT, V , MODRICH, PL , (2006) DNA MISMATCH REPAIR: FUNCTIONS AND MECHANISMS.CHEMICAL REVIEWS. VOL. 106. ISSUE 2. P. 302 -323 | 253 | 68% | 416 |
| 2 | KUNKEL, TA , ERIE, DA , (2005) DNA MISMATCH REPAIR.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 74. ISSUE . P. 681 -710 | 127 | 72% | 680 |
| 3 | KOLODNER, RD , (2016) A PERSONAL HISTORICAL VIEW OF DNA MISMATCH REPAIR WITH AN EMPHASIS ON EUKARYOTIC DNA MISMATCH REPAIR.DNA REPAIR. VOL. 38. ISSUE . P. 3 -13 | 127 | 77% | 3 |
| 4 | JIRICNY, J , (2013) POSTREPLICATIVE MISMATCH REPAIR.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 5. ISSUE 4. P. - | 126 | 72% | 65 |
| 5 | HSIEH, P , YAMANE, K , (2008) DNA MISMATCH REPAIR: MOLECULAR MECHANISM, CANCER, AND AGEING.MECHANISMS OF AGEING AND DEVELOPMENT. VOL. 129. ISSUE 7-8. P. 391-407 | 150 | 66% | 170 |
| 6 | KADYROVA, LY , KADYROV, FA , (2016) ENDONUCLEASE ACTIVITIES OF MUTL ALPHA AND ITS HOMOLOGS IN DNA MISMATCH REPAIR.DNA REPAIR. VOL. 38. ISSUE . P. 42 -49 | 112 | 79% | 3 |
| 7 | SCHOFIELD, MJ , HSIEH, P , (2003) DNA MISMATCH REPAIR: MOLECULAR MECHANISMS AND BIOLOGICAL FUNCTION.ANNUAL REVIEW OF MICROBIOLOGY. VOL. 57. ISSUE . P. 579-608 | 134 | 79% | 308 |
| 8 | HARFE, BD , JINKS-ROBERTSON, S , (2000) DNA MISMATCH REPAIR AND GENETIC INSTABILITY.ANNUAL REVIEW OF GENETICS. VOL. 34. ISSUE . P. 359 -399 | 156 | 69% | 389 |
| 9 | SPAMPINATO, CP , GOMEZ, RL , GALLES, C , LARIO, LD , (2009) FROM BACTERIA TO PLANTS: A COMPENDIUM OF MISMATCH REPAIR ASSAYS.MUTATION RESEARCH-REVIEWS IN MUTATION RESEARCH. VOL. 682. ISSUE 2-3. P. 110 -128 | 163 | 75% | 8 |
| 10 | JIRICNY, J , (2006) THE MULTIFACETED MISMATCH-REPAIR SYSTEM.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 7. ISSUE 5. P. 335-346 | 91 | 80% | 552 |
Classes with closest relation at Level 1 |